Workshop 3 + Solutions - Exploring Many Categorical Variables
- Visualising multivatiate categorical data using:
- Stacked
barplots for small problems doubledeckerandmosaicplotfor larger problems
- Stacked
You will need to install the following packages for today’s workshop:
vcdfor thedoubledeckerfunctions
install.packages("vcd")1 Multivariate categorical data
Displaying combinations of categorical variables can be quite difficult, as we can’t represent our data as simple points in a space. Instead, we summarise a categorical variable (e.g. eye colour) with multiple levels (blue, green, brown, …) by the frequencies of each of the levels in the data. When we have multiple categorical variables, we work instead with the counts of the combinations of the levels (e.g. red hair + green eyes).
All of our plots are some sort of visualisation of these counts and so are (in one way or another) variations and manipulations of stacked barplots. Multivariate categorical data is a little more complex to work with, so generally it is recommended to start with single or pairs of variables and progressively add more, rather than visualising everything all at once like a scatterplot matrix.
Download data: arthritis
The arthritis data contains the results from a
double-blind clinical trial investigating a new treatment for rheumatoid
arthritis. The variables are:
ID- patient ID.Treatment- factor indicating treatment (Placebo, Treated).Sex- factor indicating sex (Female, Male).Age- age of patient.Improved- ordered factor indicating treatment outcome (None, Some, Marked).
Treatment, Sex, and Improved
are all categorical variables. Improved is also ordinal,
since the category levels can be ordered. The question is whether the
patient improvement depends on the Treatment and Sex.
First, let’s have a quick look at the data.
head(arthritis)| ID | Treatment | Sex | Age | Improved |
|---|---|---|---|---|
| 57 | Treated | Male | 27 | Some |
| 46 | Treated | Male | 29 | None |
| 77 | Treated | Male | 30 | None |
| 17 | Treated | Male | 32 | Marked |
| 36 | Treated | Male | 46 | Marked |
| 23 | Treated | Male | 58 | Marked |
Note that each row in the data is an individual patient, rather than summaries of the counts of the different category combinations.
1.1 Mosaic plots
Mosaic plots can display the relationship between categorical variables using rectangular tiles, whose areas represent the proportion of cases for any given combination of levels. A mosaic plot of a single variable is basically a simple stacked barplot with only one bar. Looking at the patient improvement only, we see
mosaicplot(~Improved, data=arthritis,col=2:4,main='')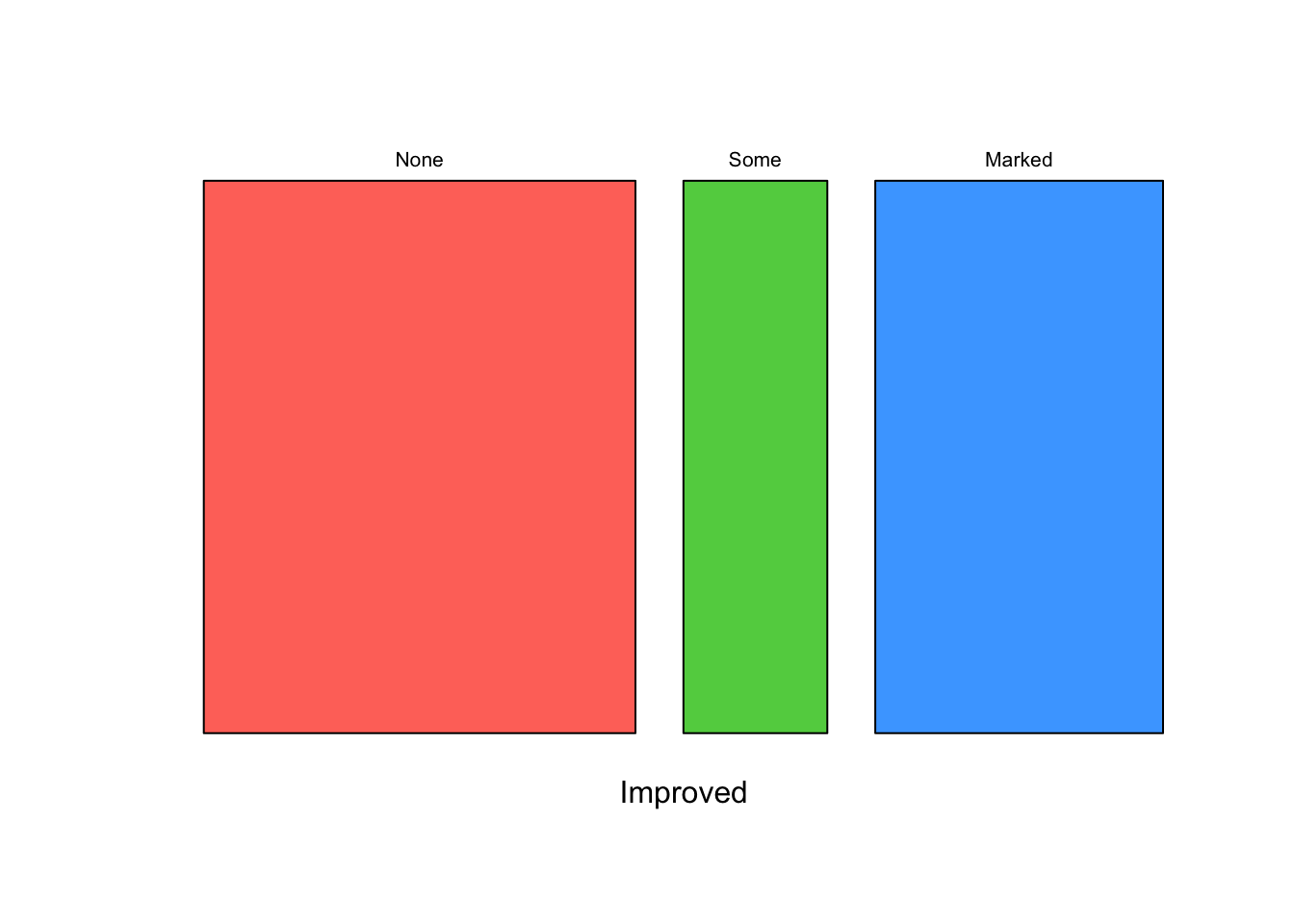
Since the None box is largest, we see that this appears
to be the most common patient outcome. But taking the Some
and Marked improvements together, it’s actually an even
split. Let’s see how this depends on what Treatment the
patient received - we would hope to see more improvement from those in
the Treated group:
mosaicplot(~Treatment+Improved, data=arthritis,col=2:4,main='')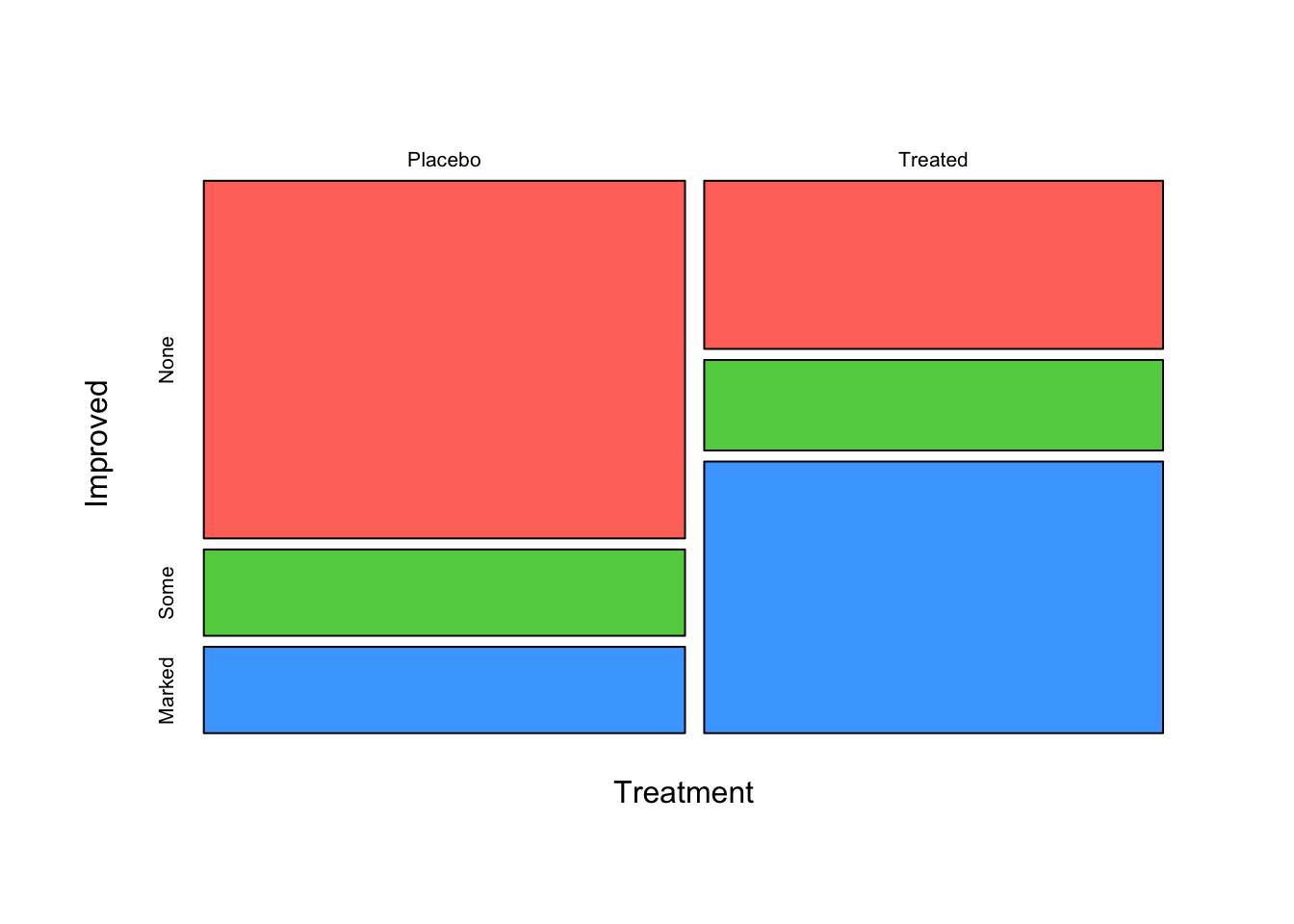
Note that the plot now has two splits: the first split is horizontal
into bars for the “Treated” and “Placebo” groups, while the second set
of splits divides each bar up into the different Improved
categories. If Treatment had no effect on the
Improved state, then we would see a regular grid
where the two sets of bars would have splits of approximately the
same size and we could draw lines from the left of the plot to the
right without cutting through any of the tiles.
However, we can see clearly that a greater proportion of patients
improved on treatment than in the placebo group and so the distribution
of Improved is different for different values of
Treatment, which may point towards an association between
these variables and maybe hints at treatment being effective!
If we reverse the positions of Improved and
Treatment in the function, it change the order in which the
bars are split:
mosaicplot(~Improved+Treatment, data=arthritis, col=2:4,main='')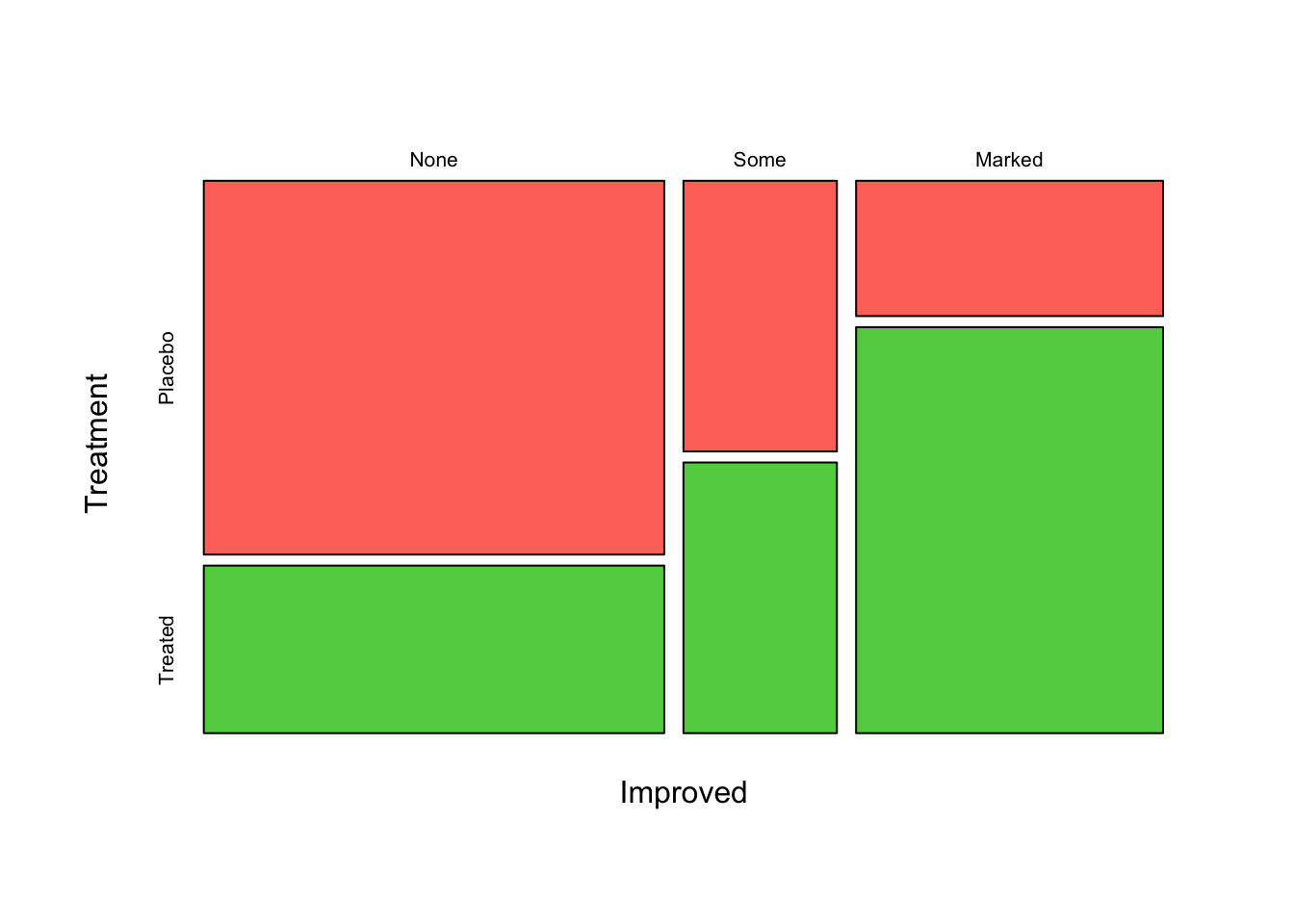
Now we have three vertical bars for Improved, each
sub-divided into the Treatment groups. This plot now shows
how the patients with different Improved levels break down
into the Treatment groups. So, we would read this as saying
for those patients with a Marked improvement, the majority were Treated
rather than given Placebo. Usually, it is best to split on the response
variable at the end, as we had done in the previous plot.
1.2 More variables
We can continue to add variables to the plot and break down the
results into more groups. For instance, we can introduce
Sex as a variable
mosaicplot(~Treatment+Sex+Improved, data=arthritis,col=2:4,main='')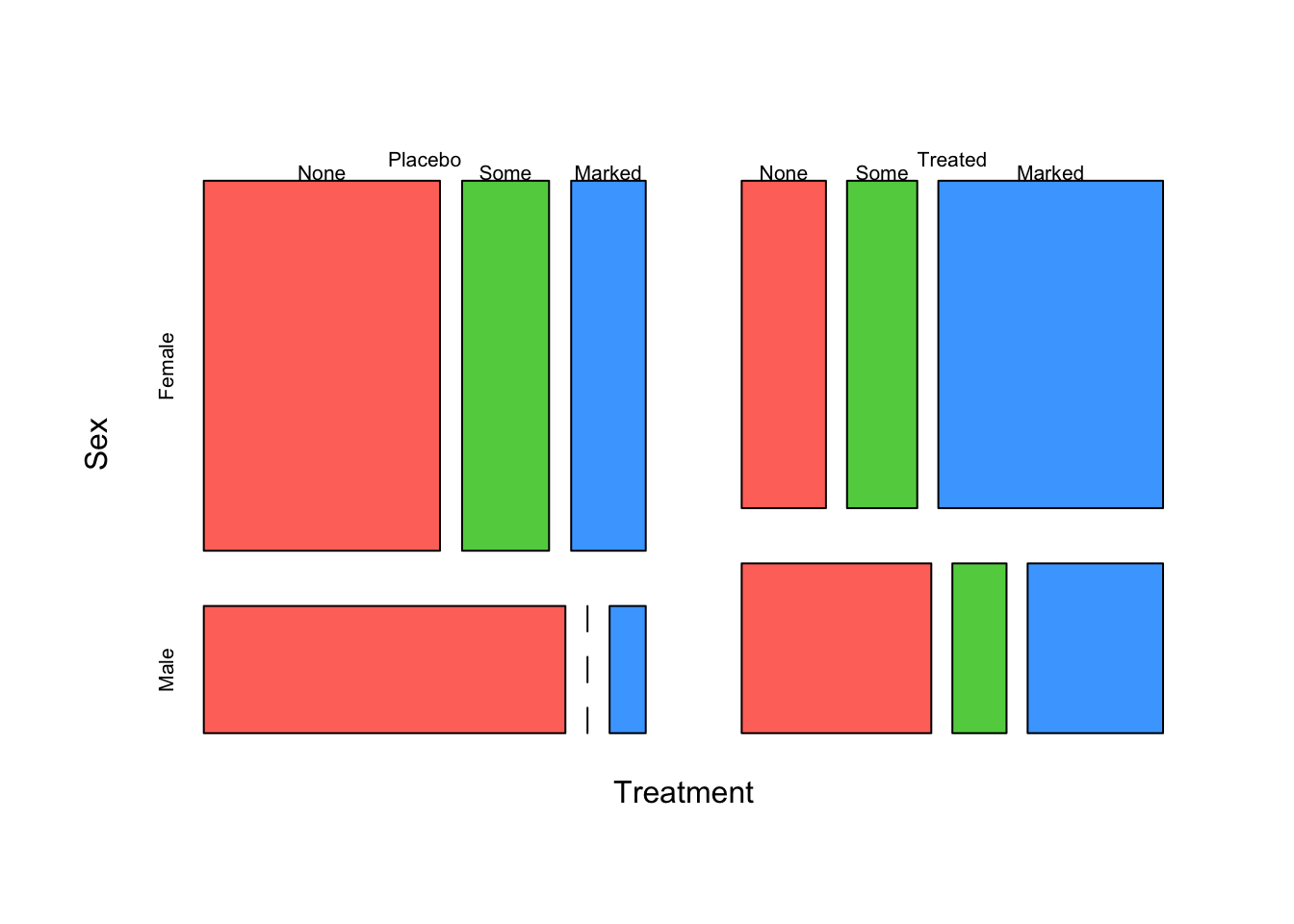
Here we can see:
- The Treatment groups look to be roughly equal in size
- There are fewer men than women in the study (the
Malerows are narrower than theFemale) - There are no
Malepatients in thePlacebogroup who only hadSomeimprovement - this is indicated by the dashed line where this bar should be. Treatmentappears to have a positive effectFemalepatients seemed to improve the most in general, and particularly underTreatment
It is often worth reordering the variables in the mosaic plot formula
to see if a different sequence of splits is a more effective
visualisation for your problem. Generally, our dependent variable of
interest is split last following the ~ in the function
call.
- Experiment with the ordering of the variables in the call to the
mosaicplotfunction to see how different orderings produce different presentations of the data.
## there are 3!=6 different orderings - here are a couple
mosaicplot(~Improved+Sex+Treatment, data=arthritis,col=2:4,main='')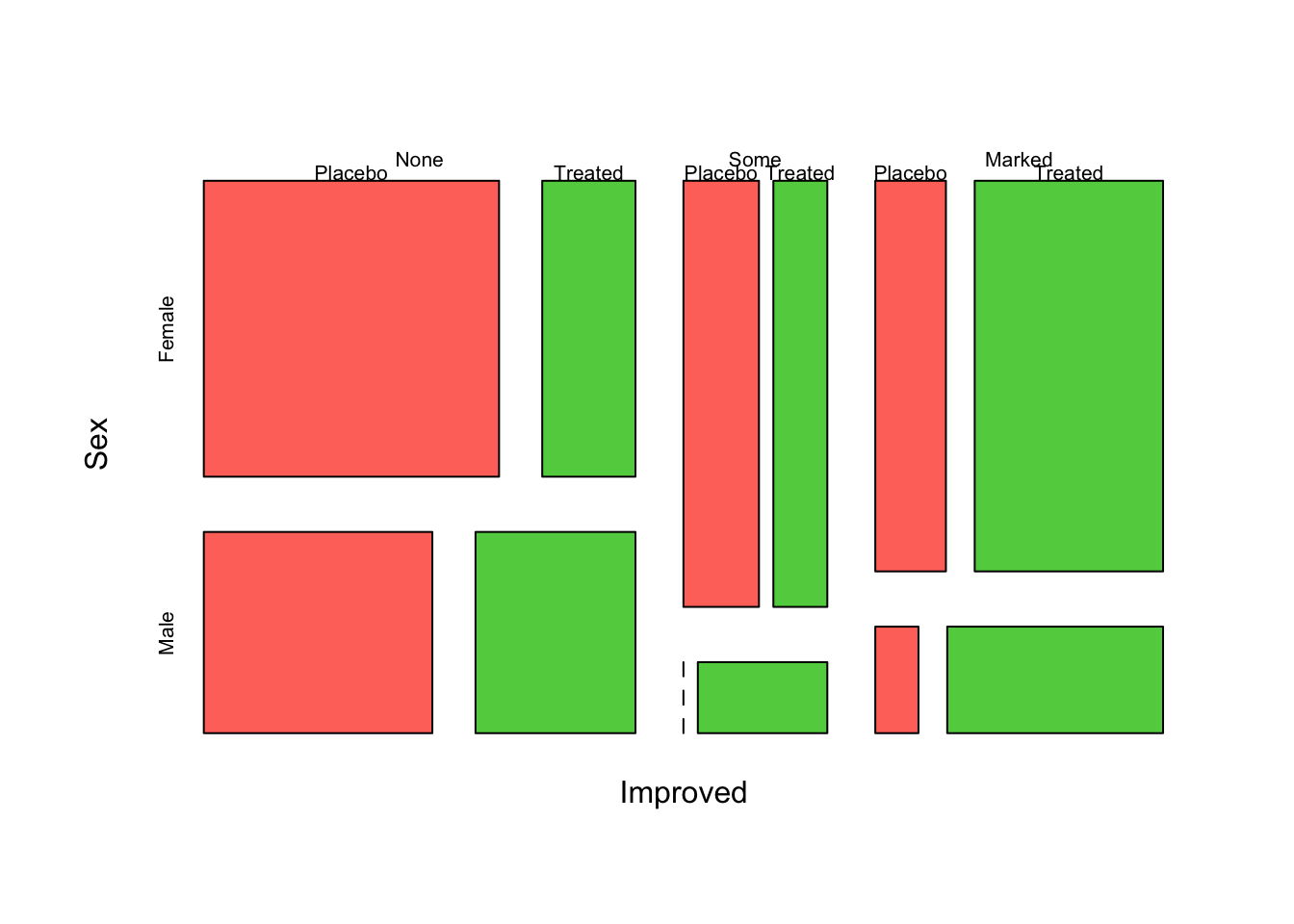
mosaicplot(~Sex+Improved+Treatment, data=arthritis,col=2:4,main='')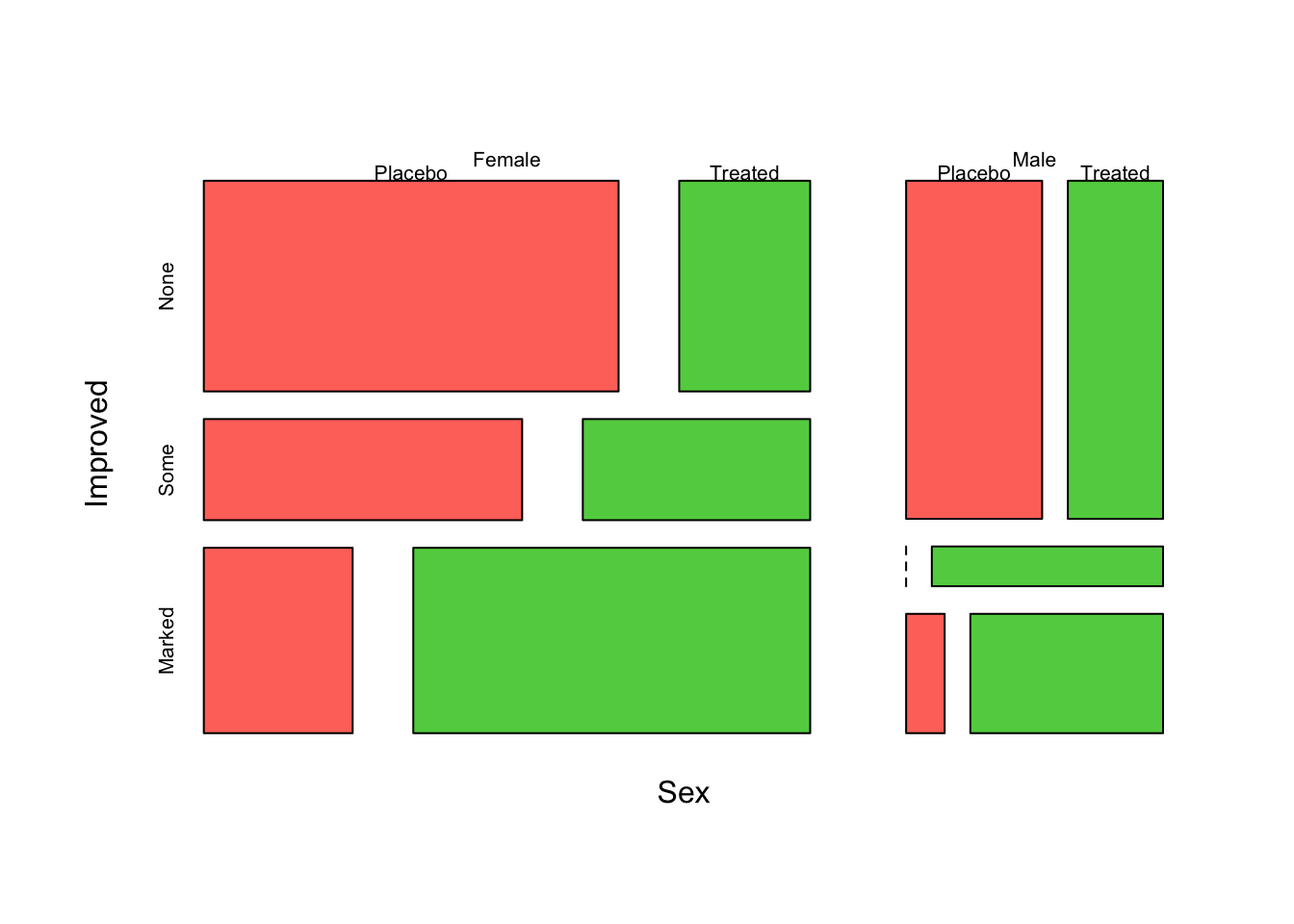
1.2.1 Directions of the splits
Note that in the previous example, the mosaic plot divides the plot using different directions depending on the order the variables are specified:
Treatment- first split, verticalSex- second split, horizontalImproved- final split, vertical
A doubledecker plot is a version of a mosiac plot where
all of the splits in the data are vertical, except for the last one.
This effectively produces a type of stacked barplot, which we can
achieve by setting the direction argument as follows:
mosaicplot(~Treatment+Sex+Improved, data=arthritis, dir = c("v", "v", "h"),col=2:4)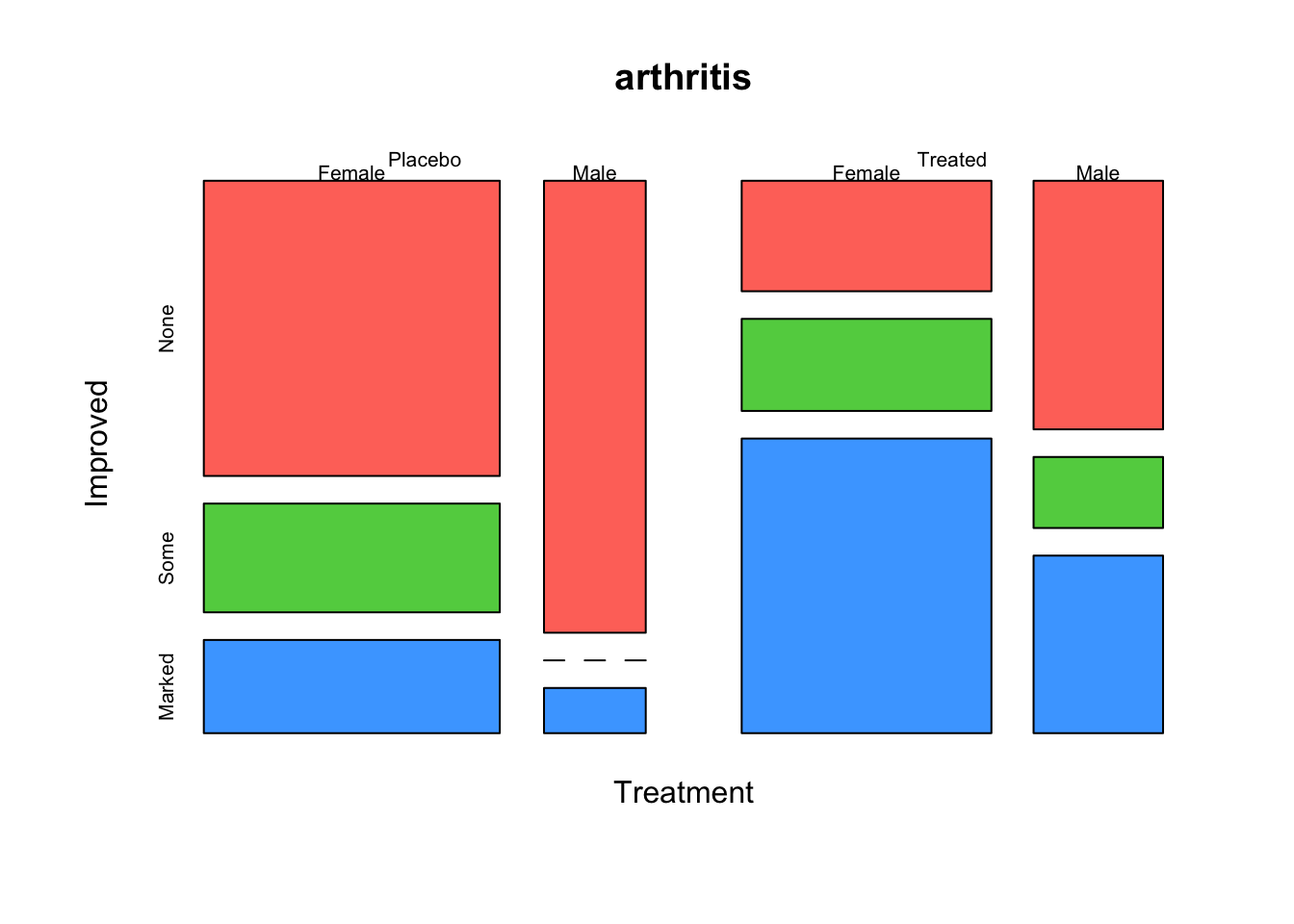
Or, using the doubledecker function directly gives an
almost identical plot:
doubledecker(Improved~Treatment+Sex, data=arthritis)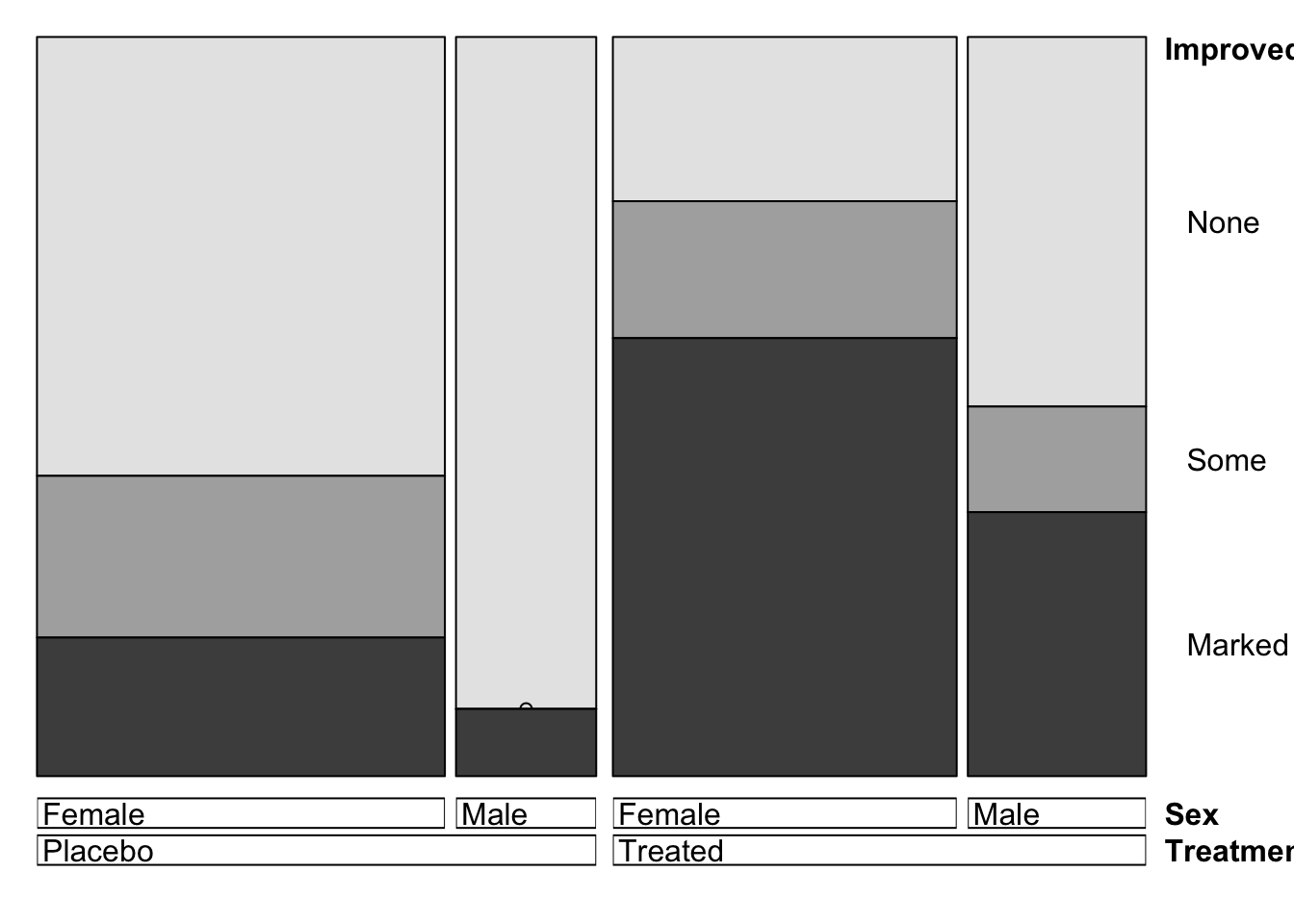
1.2.2 Adding some colour
The default plots are somewhat dull and monochrome. Note that the
different shadings used in the plot correspond to the different levels
of the last cut variable, i.e. the dependent variable which is
Improved here. So, if we supply one colour for each level
of that variable we get the following plot:
mosaicplot(~Treatment+Sex+Improved, data=arthritis, main='', col=c("wheat", "cornflowerblue", "tomato1"))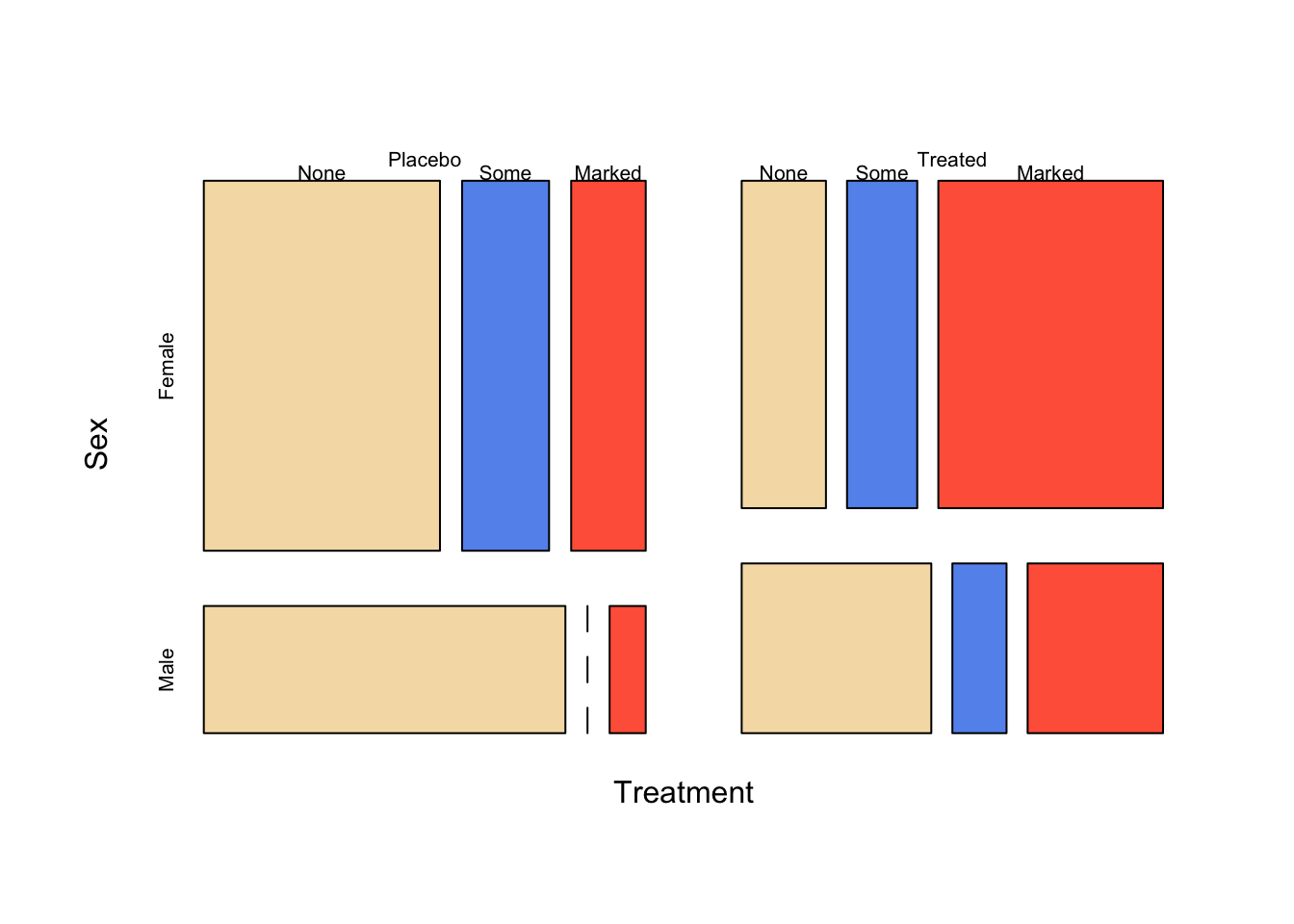
Note: In general, setting colours on mosaic plots can be quite fiddly. We won’t make much use of it, apart from an automatic coloring technique that we describe below.
1.3 Using colour to highlight unexpected patterns
A different but helpful use of colour is the shade=TRUE
option. There is a formal
statistical test to assess if two (or more) categorical variables
are independent (i.e. have no association). This test works by comparing
our observed data with what we would expect to see from a similar
problem under this independence hypothesis. The shade=TRUE
option works by colouring any tiles in the mosaic that are in
disagreement with that hypothesis. This can help us identify any
combinations that are unusually common or rate:
mosaicplot(~Treatment+Sex+Improved, data=arthritis,shade=TRUE,main='')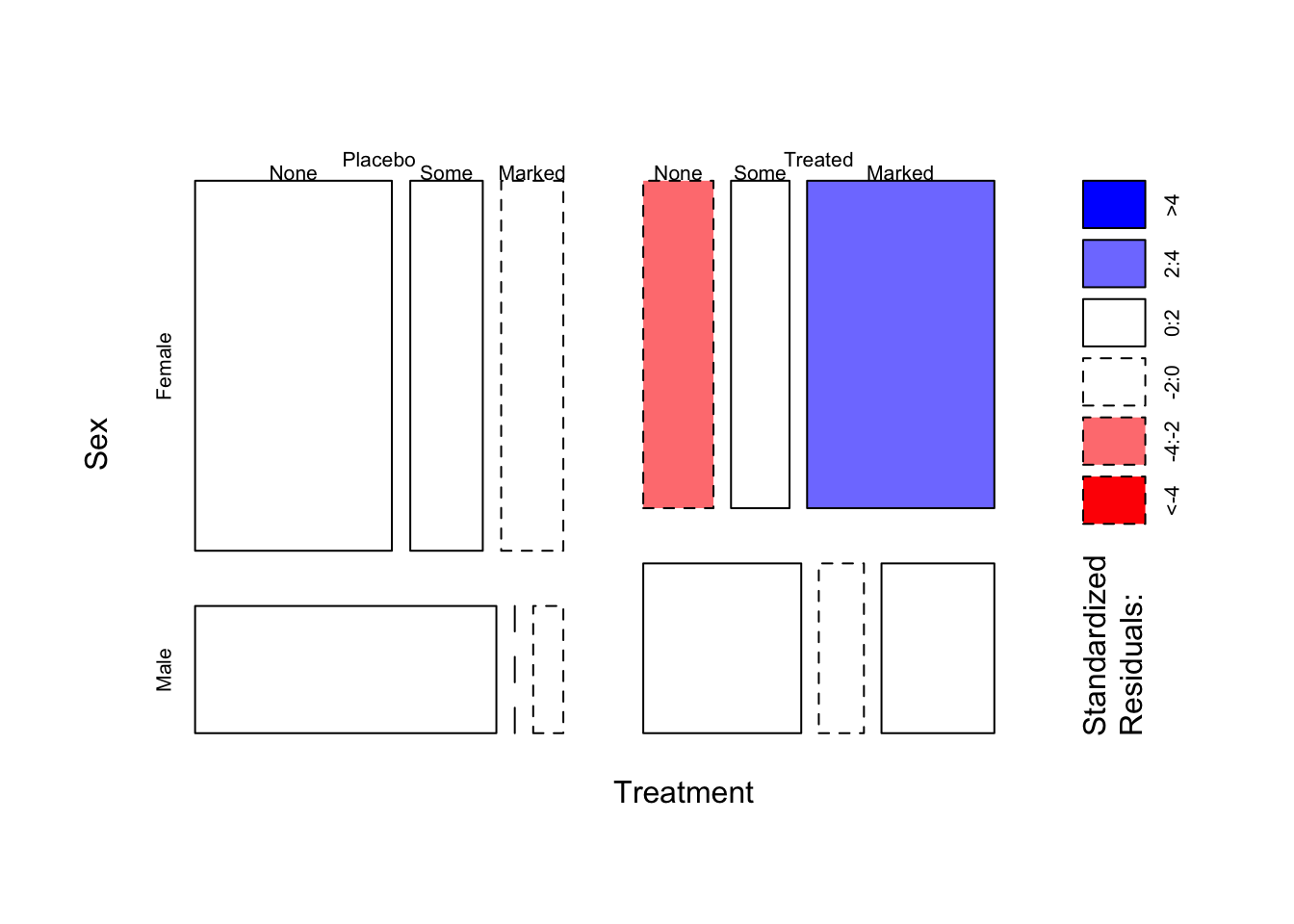
Tiles are shaded blue when more cases are observed than expected
given independence, and shaded red when there are fewer cases than
expected under independence. The strength of colour indicates how
“surprising” those values are. The plot here is showing that most of the
variation is not significant (coloured white), but in the
Female and Treated group there is a
surprisingly high number of patients who display a Marked
improvement (blue-ish) - and consequently, fewer than expected (red-ish)
whose improvement was None.
- Use the
mosaicplotfunction withshadeand thedirectionarguments to create a “doubledecker” version of the mosaic plot above.
mosaicplot(~Treatment+Sex+Improved, data=arthritis,shade=TRUE,main='',dir = c("v", "v", "h"))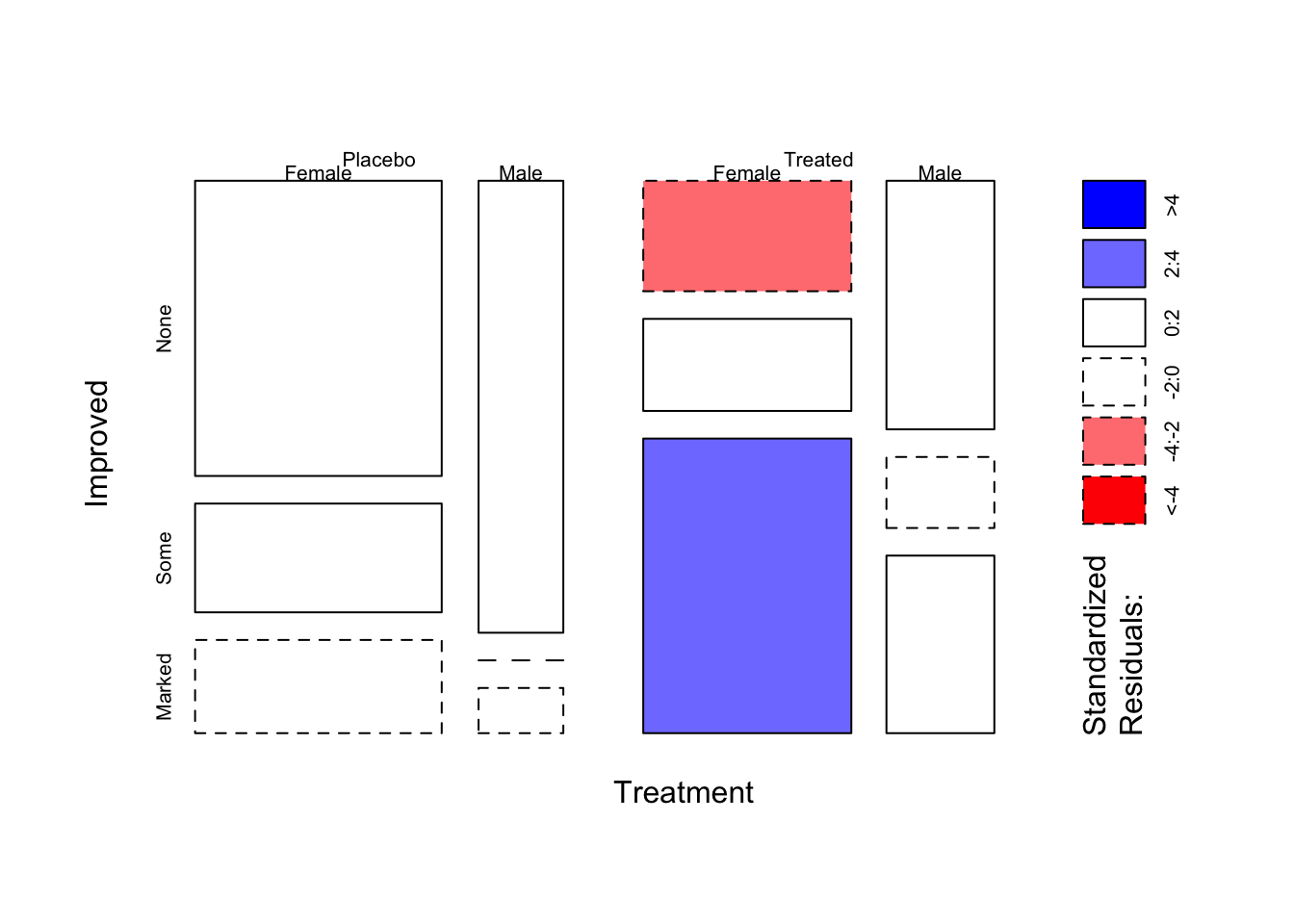
1.4 Data set 3: Alligators
Download data: alligator
The alligator data, from Agresti (2002), comes from a
study of the primary food choices of alligators in four Florida lakes.
The goal is to try and learn something about the food choice of the
different alligators. The variables are:
lake- one of four lakes:George,Hancock,Oklawaha, andTraffordsex-maleorfemalesize-smallorlargefood- the food preferences of the alligators in five categories:fish,invertebrates,reptile,birdandother.
As usual, we begin with a quick look at the data to see what we’re dealing with:
head(alligator)| lake | sex | size | food | count |
|---|---|---|---|---|
| Hancock | male | small | fish | 7 |
| Hancock | male | small | invert | 1 |
| Hancock | male | small | reptile | 0 |
| Hancock | male | small | bird | 0 |
| Hancock | male | small | other | 5 |
| Hancock | male | large | fish | 4 |
Here, unlike the arthritis data, each row does not
represent an individual alligator but all of the alligators found with
the given combinations of categorical variables. So, for example, we
have seen 7 alligators with attributes (Hancock,
male, small, fish). This is a
slightly different format than we saw above, so we’ll need to deal with
it slightly differently.
To produce the counts needed for our plots, we need to use the
cross-tabulation function xtabs that we used with our
barplots. So, to generate the counts of alligators in each
lake, we first compute
xtabs(count~lake, data=alligator)## lake
## George Hancock Oklawaha Trafford
## 63 55 48 53and then pass this to our mosaic function for
plotting:
mosaicplot(xtabs(count~lake, data=alligator),col=2:5)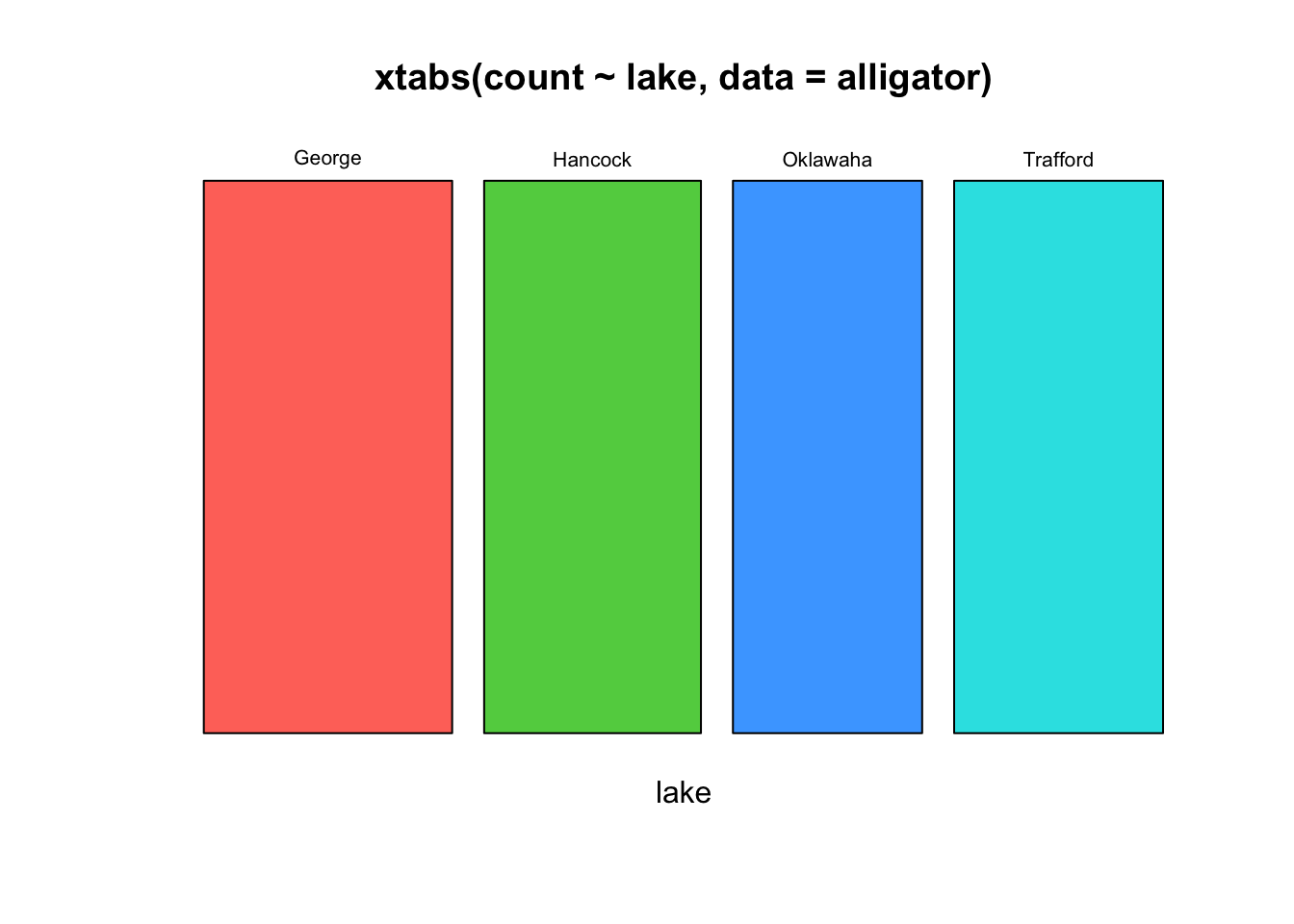
Alternatively, with a single variable we could just draw a
barplot, which is probably a little easier to read!
barplot(xtabs(count~lake, data=alligator),col=2:5)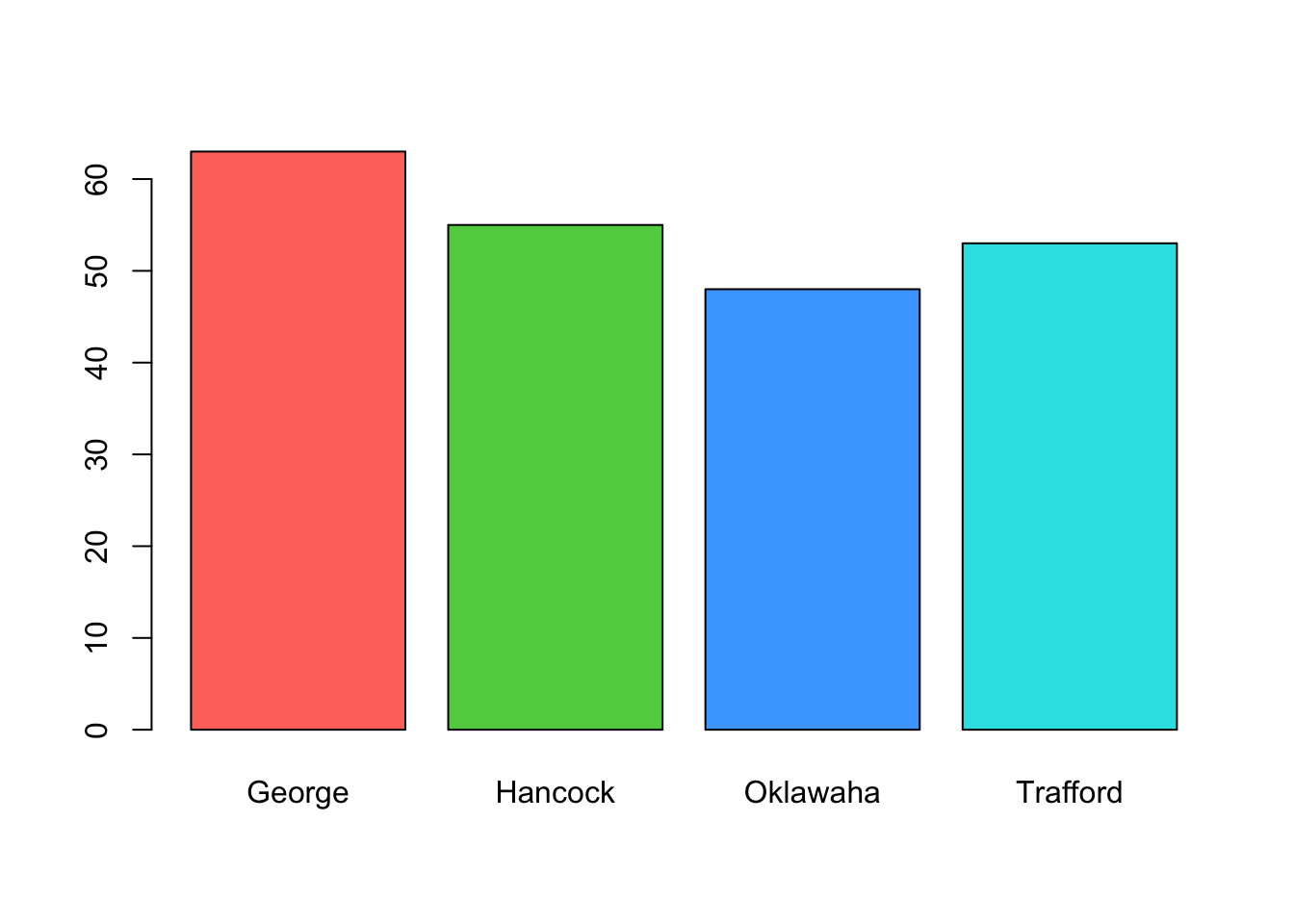 Note that the mosaicplot is showing the proportions in the different
lakes by the width of the bars, whereas the barplot uses the height. We
see there are slight differences between the numbers of alligators
observed in the different lakes, but they don’t appear to be
substantial.
Note that the mosaicplot is showing the proportions in the different
lakes by the width of the bars, whereas the barplot uses the height. We
see there are slight differences between the numbers of alligators
observed in the different lakes, but they don’t appear to be
substantial.
Note that the only difference with working with these data (which
include the counts as a variable) and the previous data set (which did
not include the counts) is that we must do the aggregating of the data
in the xtabs function first, instead of directly in
mosaicplot. The syntax and formula for splitting the data
is the same.
- Now investigate the distributions of the other categorical variables
individually:
sex,size, andfood. You can use whatever plot you prefer. Try and answer the following questions:- Are the sexes of alligators evenly distributed?
- What about the different sizes?
- Which food type is most popular?
mosaicplot(xtabs(count~sex, data=alligator))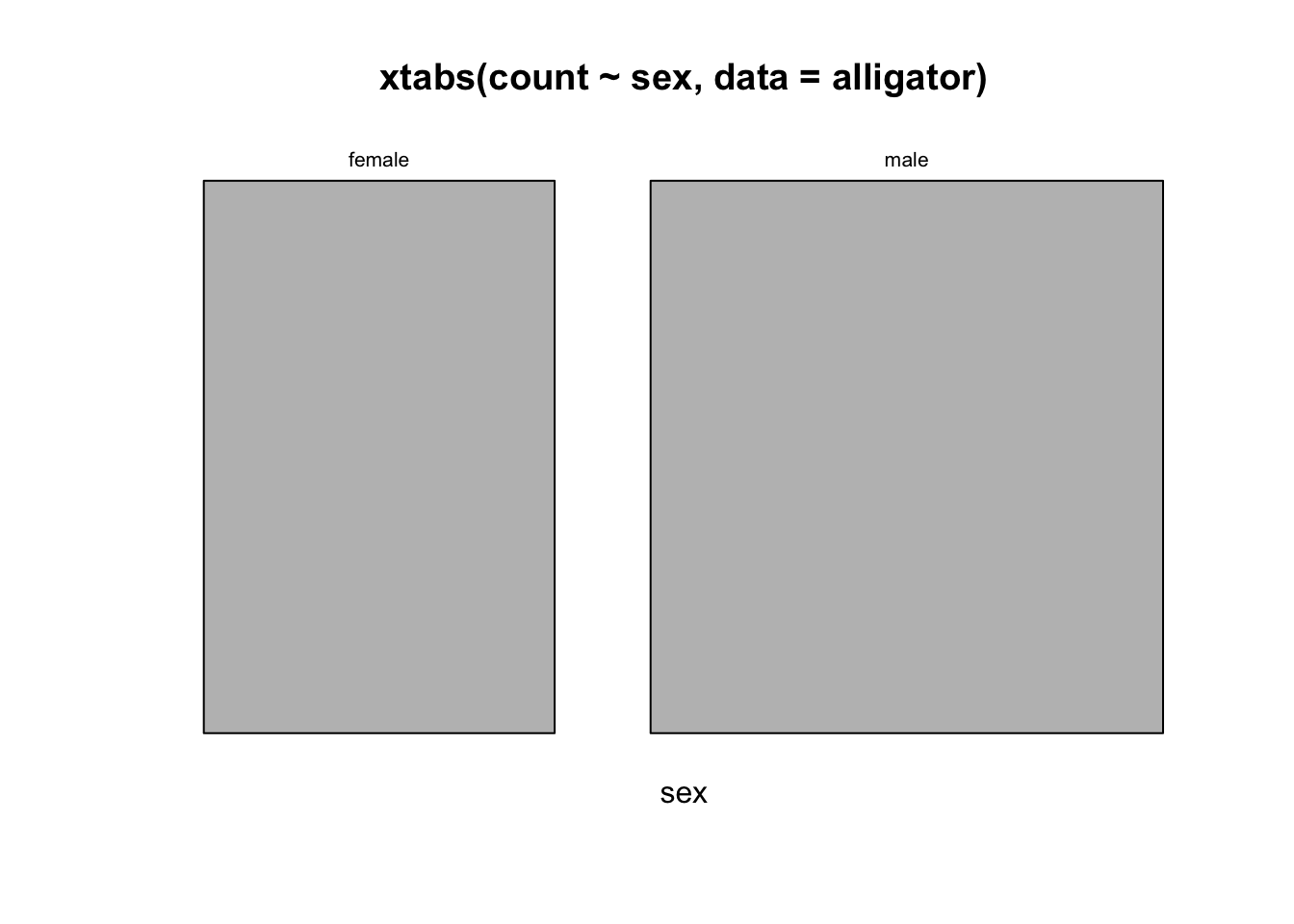
## more m than fmosaicplot(xtabs(count~size, data=alligator))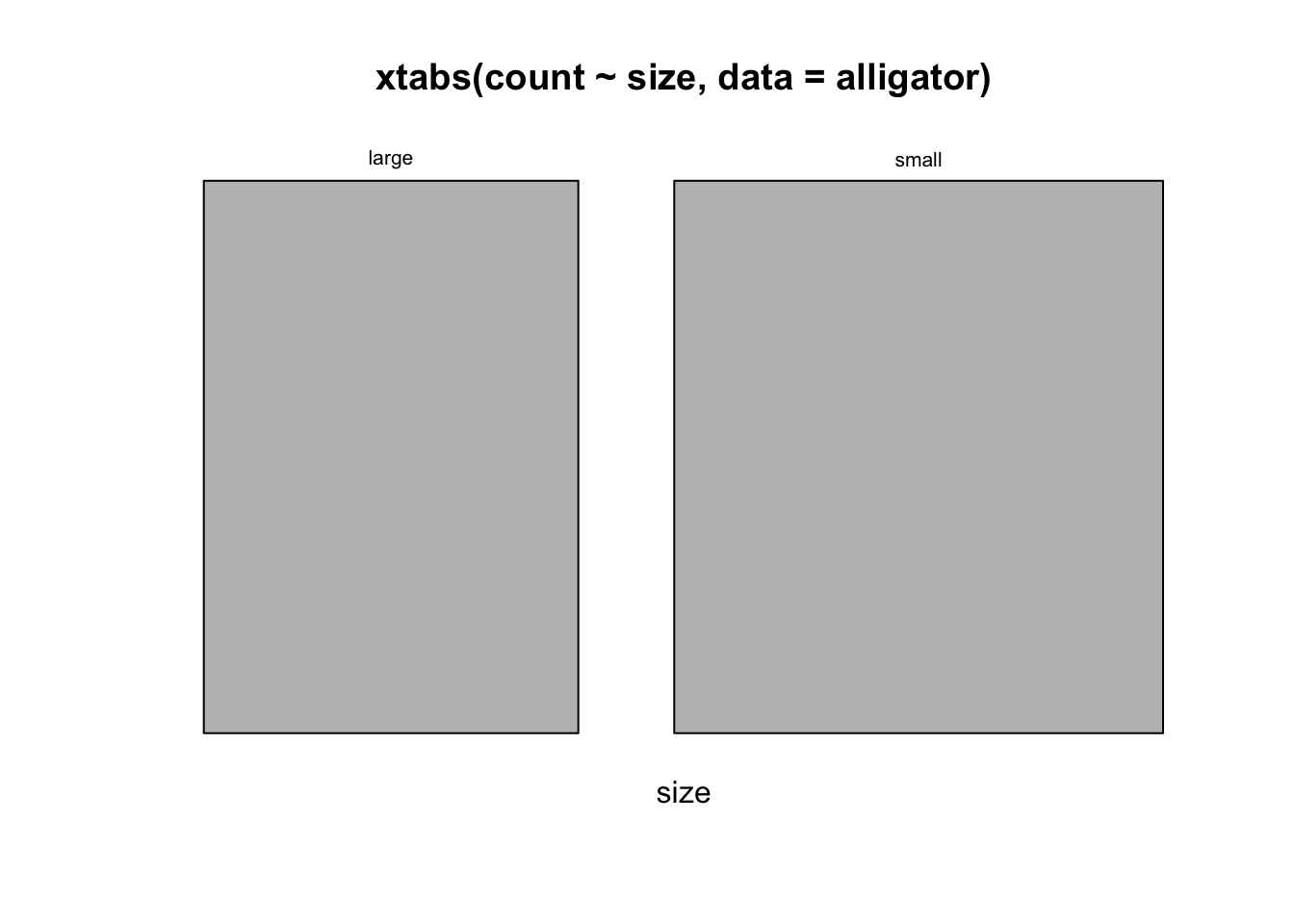
## slightly more small than largemosaicplot(xtabs(count~food, data=alligator))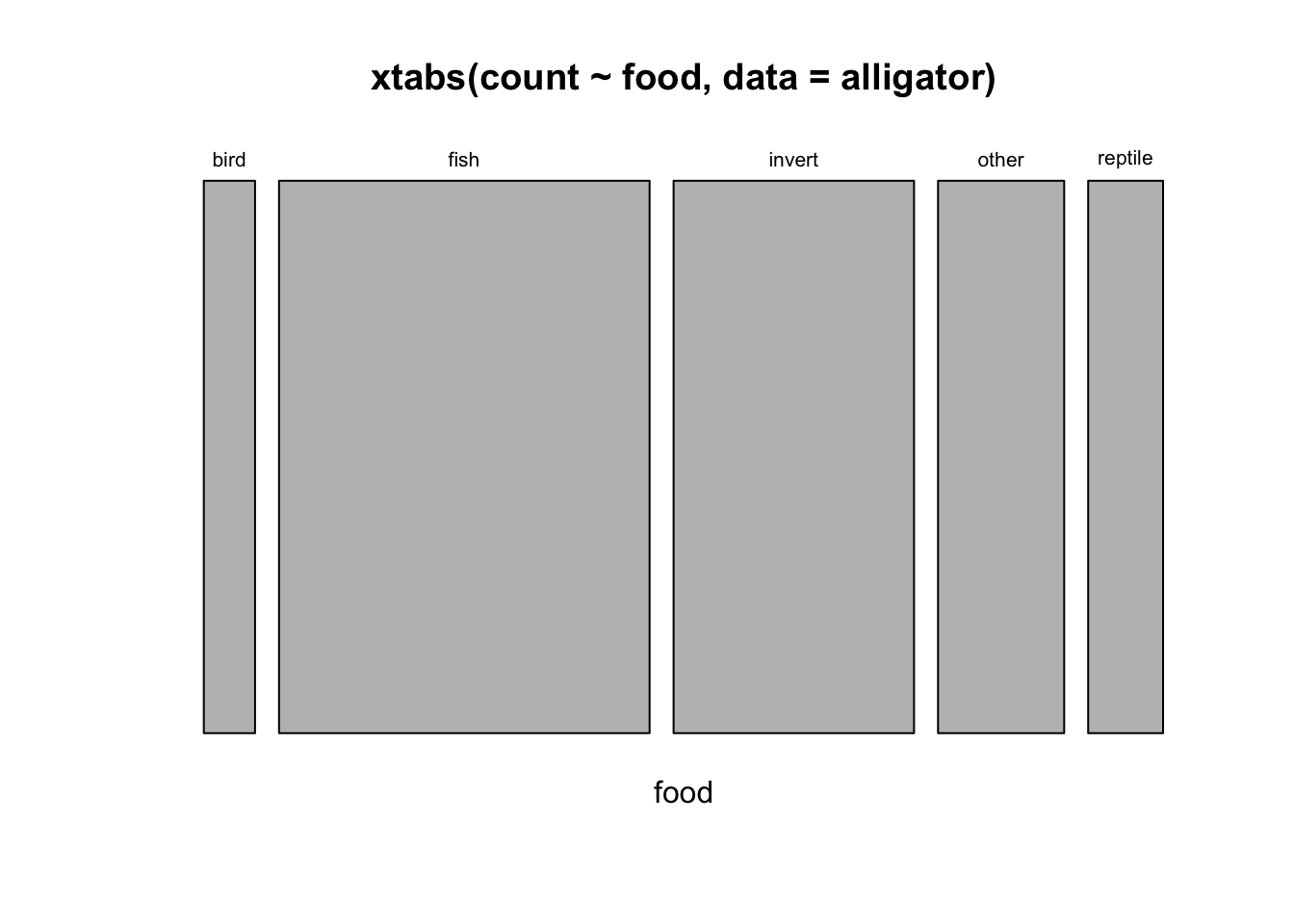
## most eat fish, then invertebrates1.5 More than two variables
The strength of mosaic plots is when considering the combination of multiple categorical variables at once. To keep things manageable, let’s looks at some potentially interesting pairs of variables first:- Use
mosaicplotto visualise thesizeandsexvariables together. Remember, if there is no association here then we would expect a regular grid. What associations do you find? - What about
sizeandfood? - Draw the
doubledeckerplots of the same variables - how do they compare to the mosaic plot?
mosaicplot(xtabs(count~size+sex, data=alligator))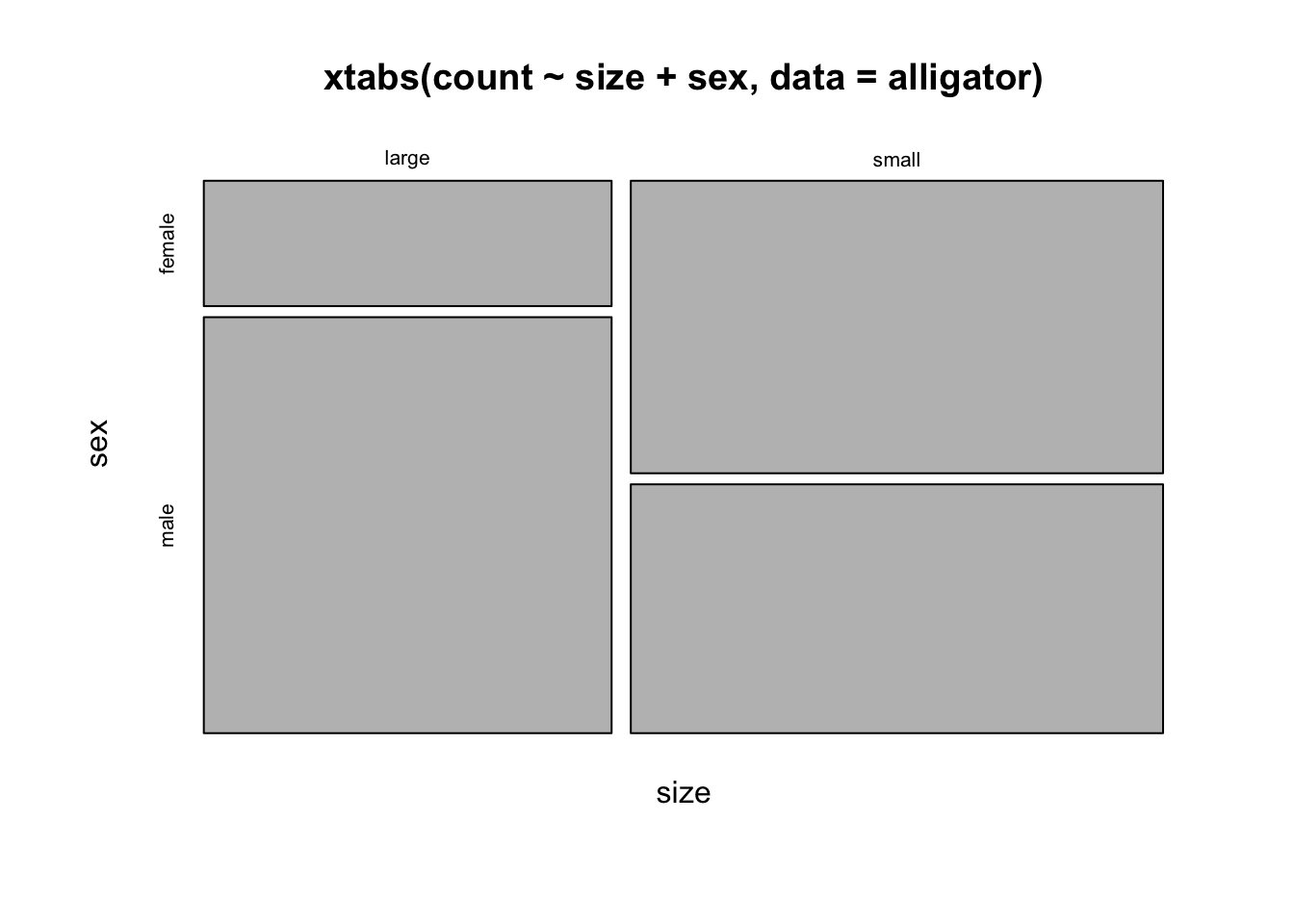
doubledecker(xtabs(count~size+sex, data=alligator))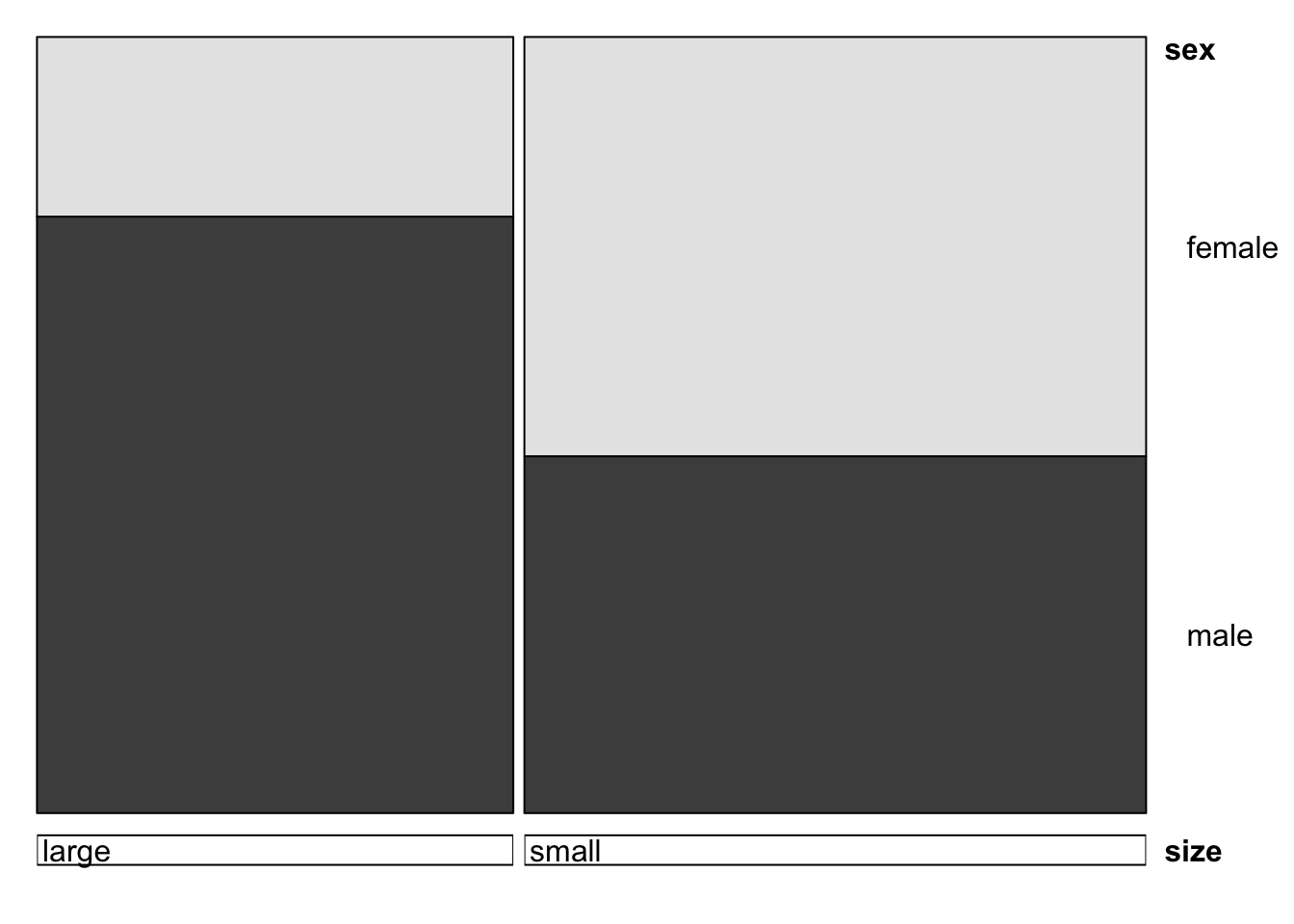
## more males are larger, females are smallermosaicplot(xtabs(count~size+food, data=alligator))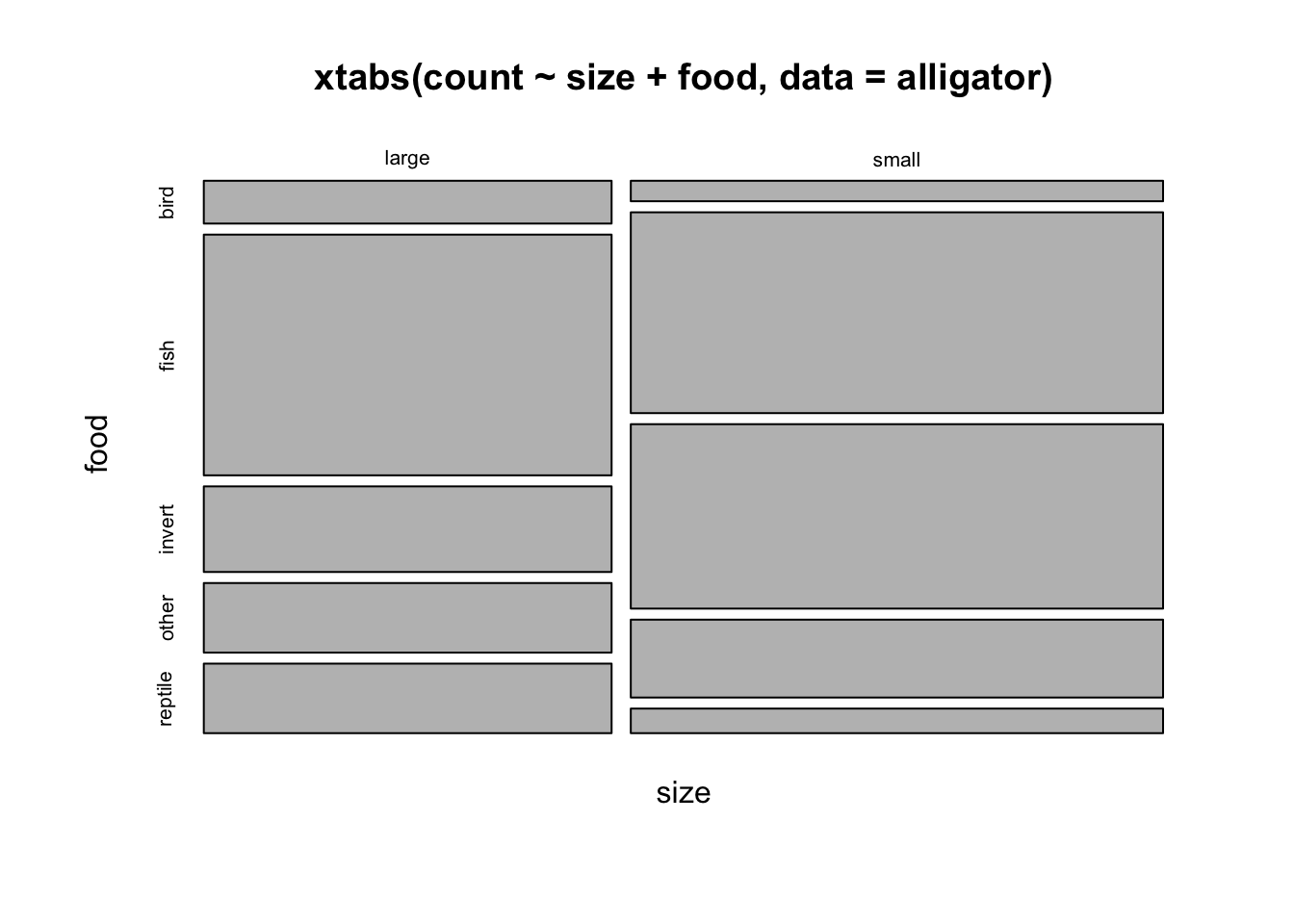
doubledecker(xtabs(count~size+food, data=alligator))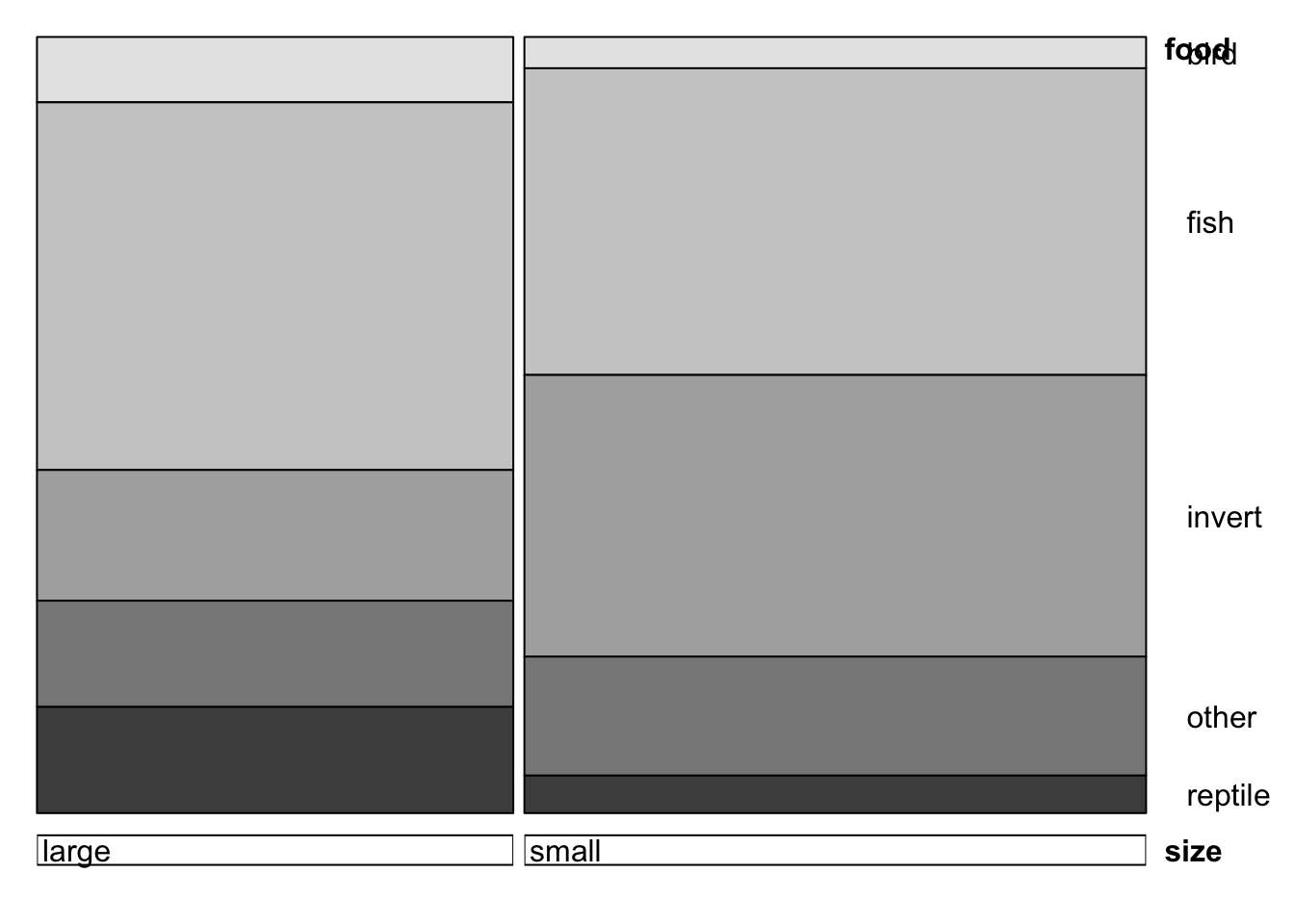
## big alligators eat more fish+reptile, and less invertebratesWe can even make a matrix of all the two-way mosaic plots in the style of a scatterplot matrix by the following command:
pairs(xtabs(count~.,data=alligator))Using a . on the right side of the formula is a
shorthand for “include everything”.
- Can you locate the plots of
sizeandsex, andsizeandfoodwithin the matrix? - Do you see any other potential associations (or lack of associations) here? Remember, “no association” will mean the mosaic is divided into an approximately regular grid.
## size:sex plot is in position (1,3), size:food is in (3,4)
## seem to be a fair few associations here, except for food:sex
## take a closer look:
mosaicplot(xtabs(count~sex+food, data=alligator))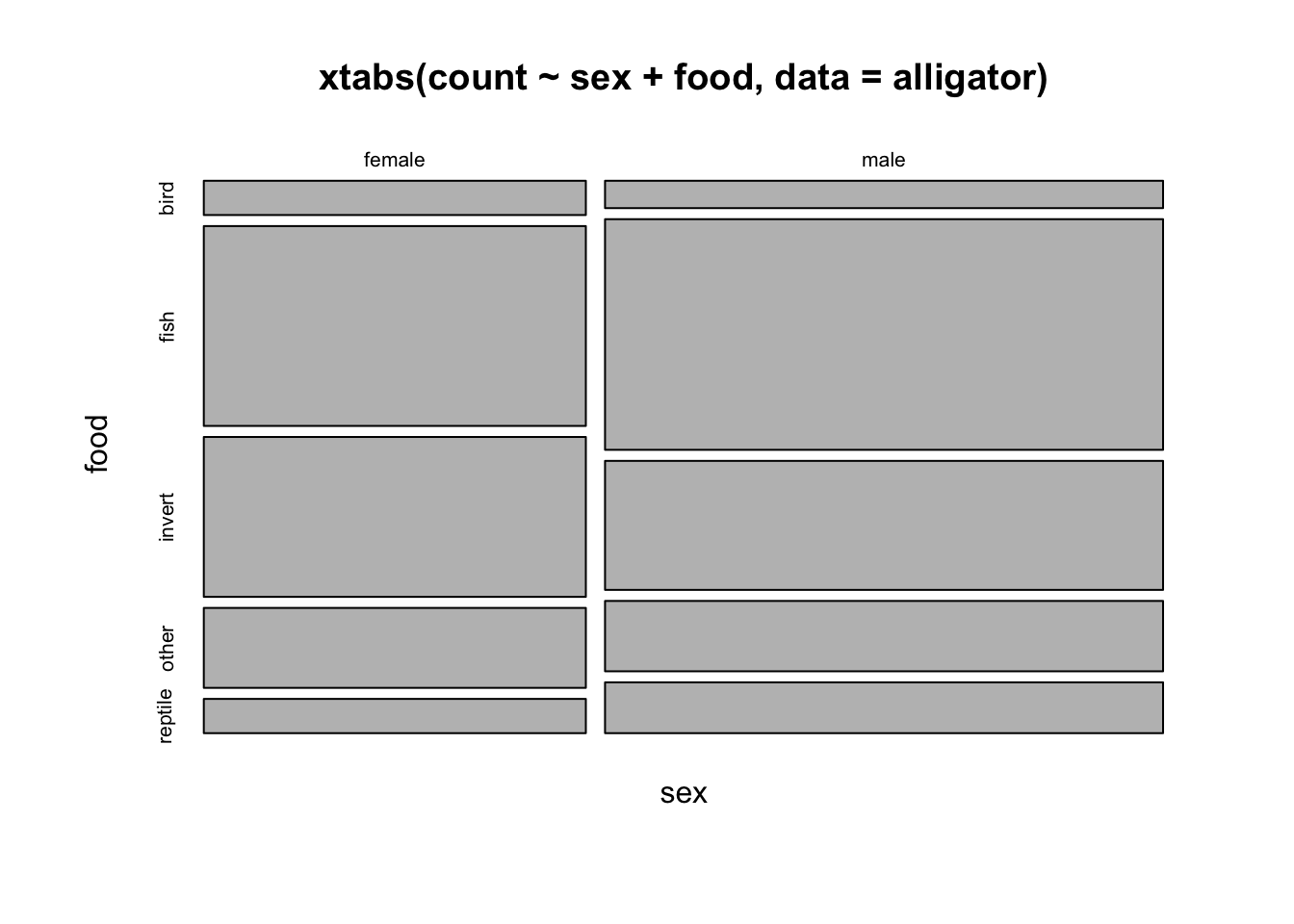
## looks pretty regular, no obvious surprisingly large/small blocks.
## Let's try shading the big plot for more emphasis
pairs(xtabs(count~.,data=alligator), shade=TRUE)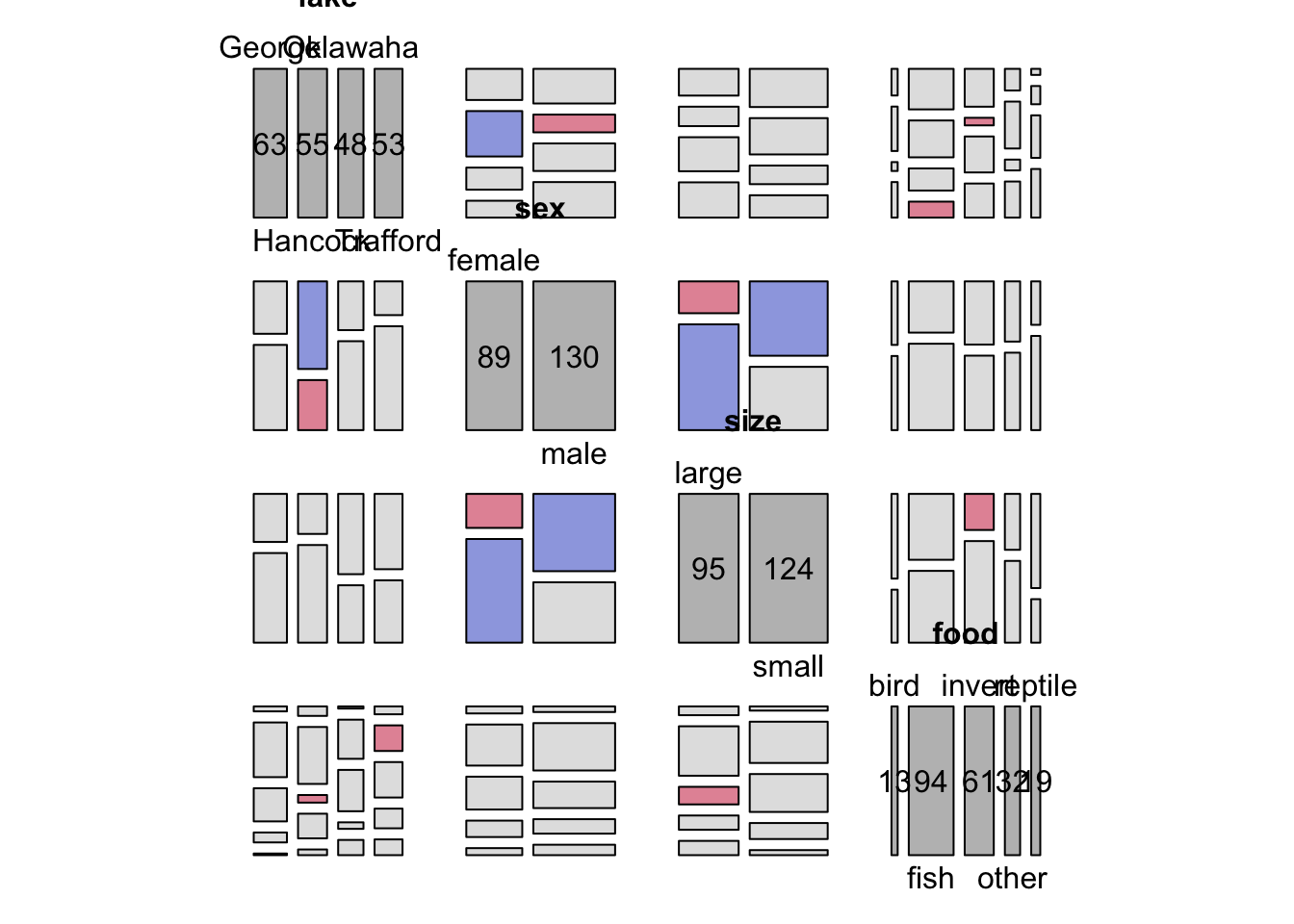
size, sex and food all
together.
- A doubleddecker plot is often more readable at first. Make a
doubledecker plot of
food,sizeandsex- order the variables so that each bar is split into sections according to thefood. - Now try the mosaic plot and use the
shade=TRUEoption. Try and achieve the same ordering so that food is the final split. What combinations have been highlighted, and how would you interpret them? - Do you see any other potentially interesting features here?
doubledecker(xtabs(count~size+sex+food, data=alligator))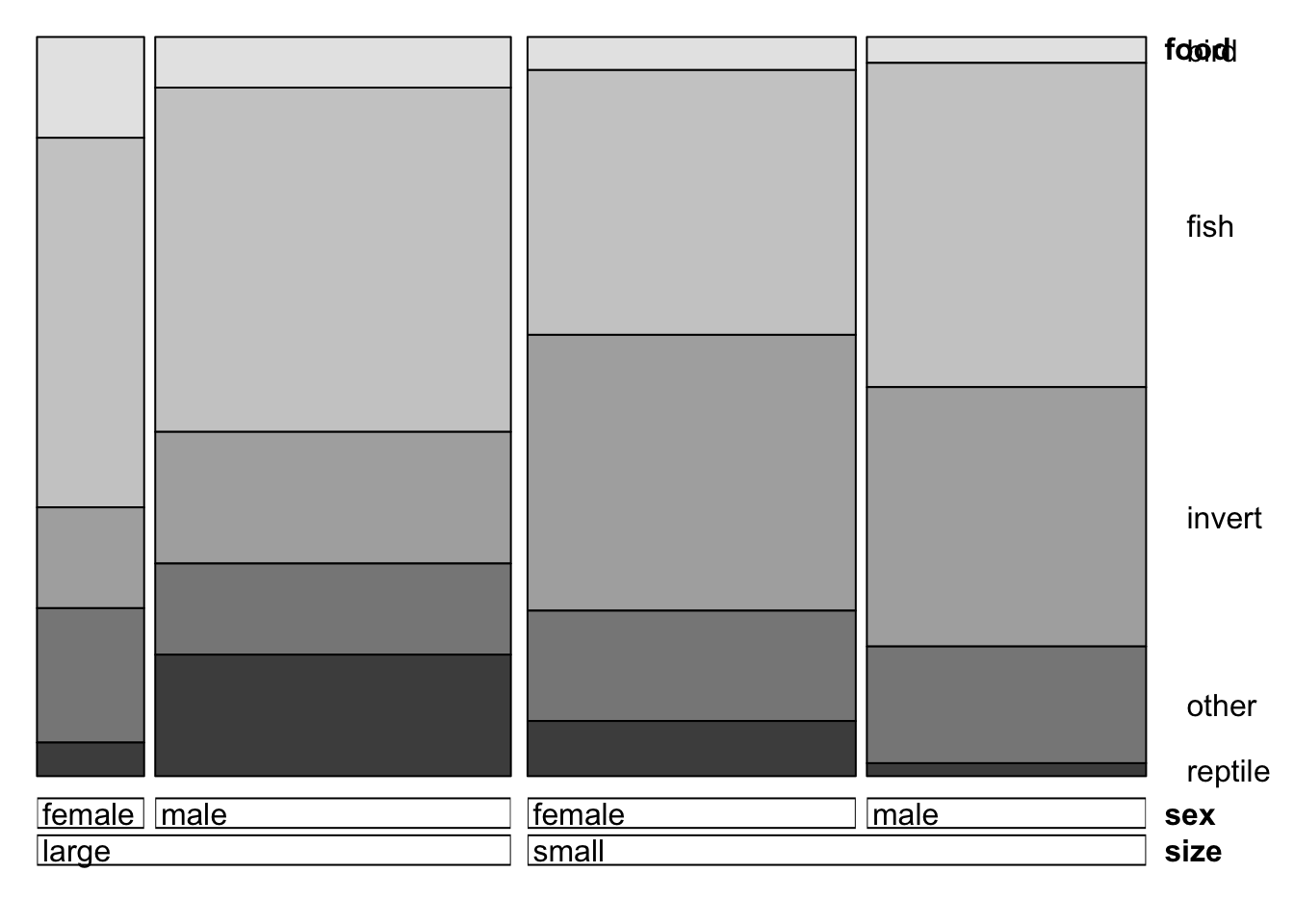
mosaicplot(xtabs(count~size+sex+food, data=alligator),shade=TRUE)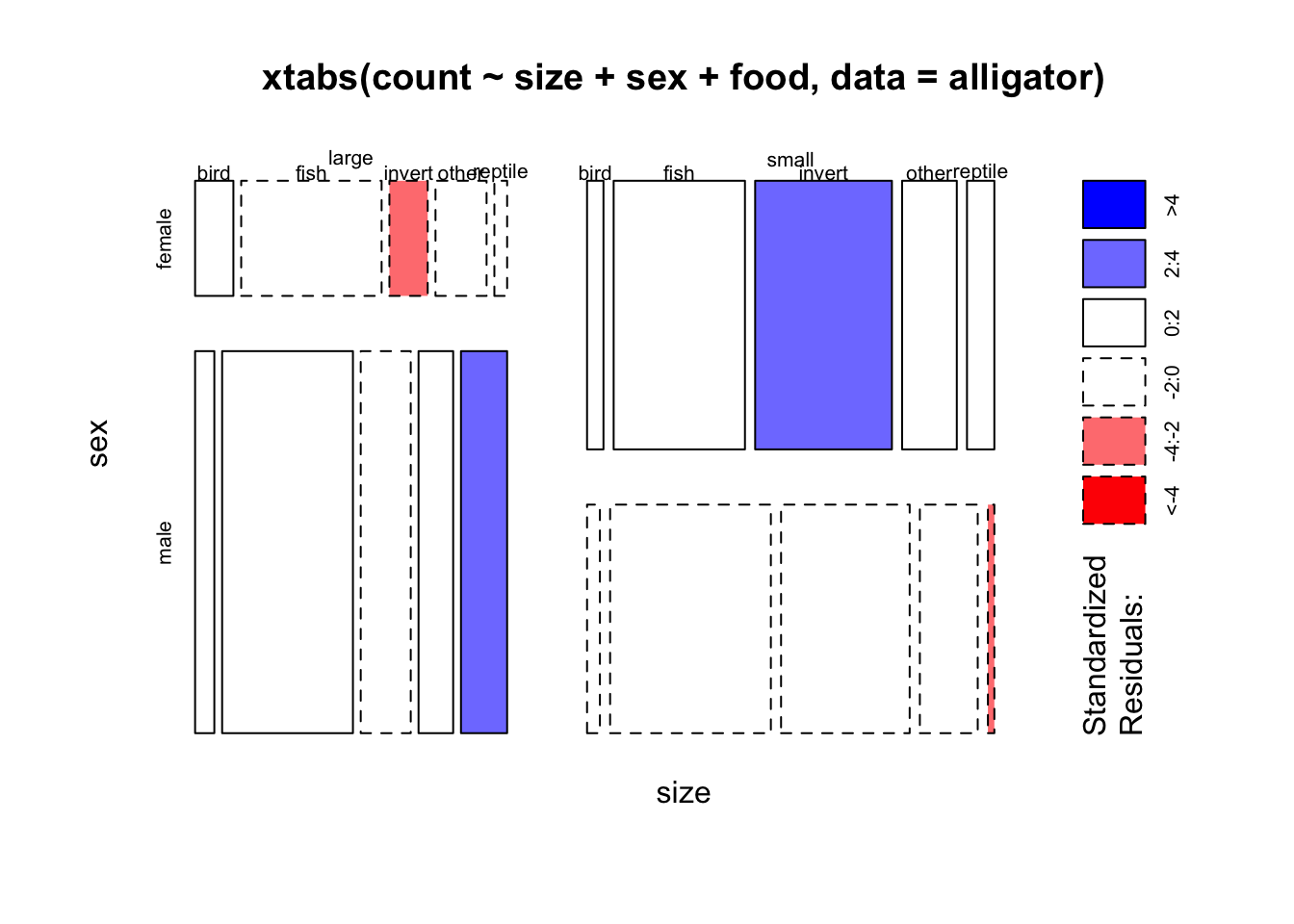
## large males eat more reptiles, small males eat surprisingly few
## (are they eating each other...?)
## small females eat more invertebrates, large females eat few## the four-way table goes a bit mad...
mosaicplot(xtabs(count~size+sex+food+lake, data=alligator),shade=TRUE)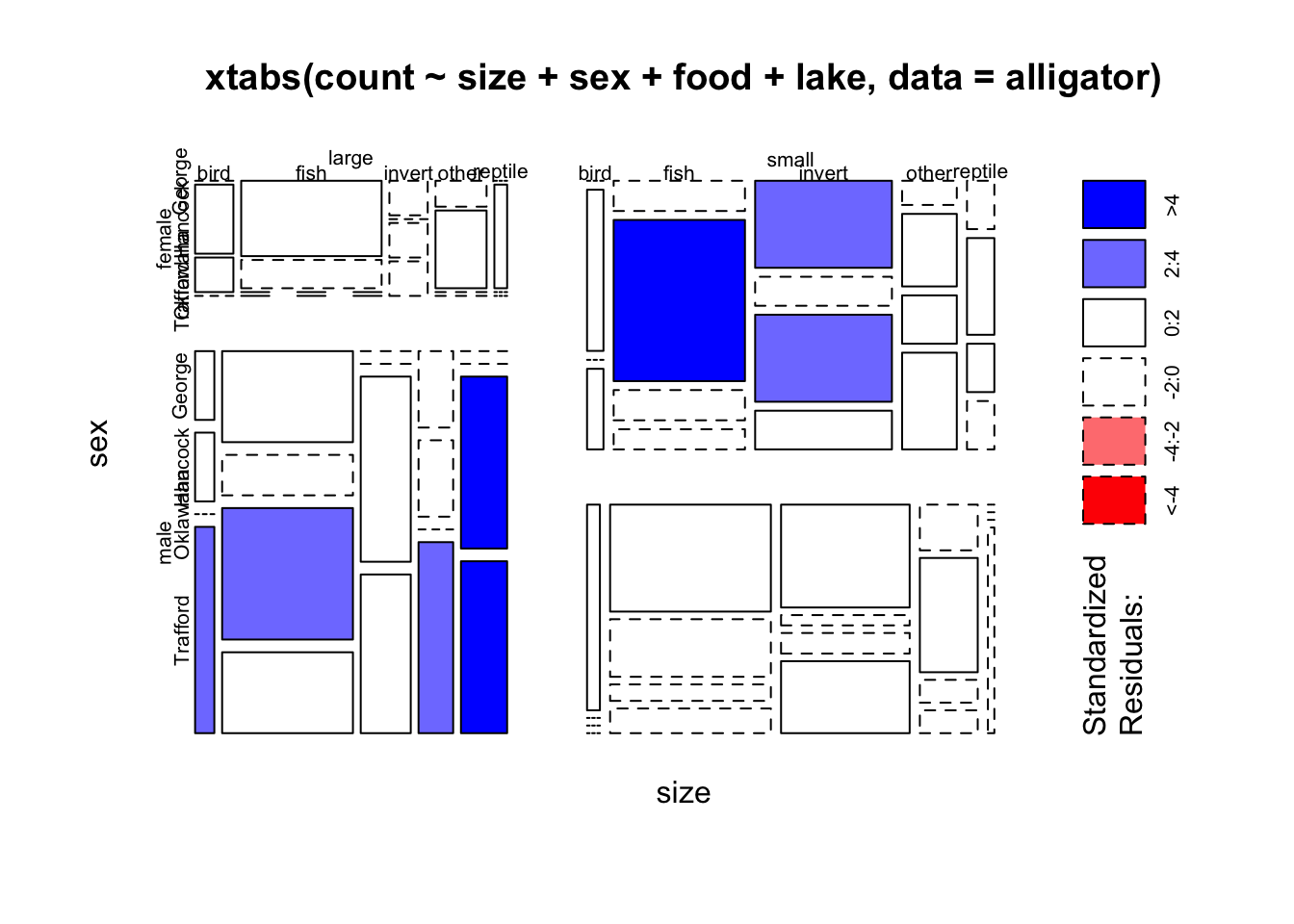
## but does reveal some features that may be worth digging into, though its
## become very hard to digest2 Variations on Standard Plots
2.1 Grouped and stacked barplots
Barplots can be used effectively to display combinations of categorical variables. However, they require a little more setup to provide the data in the correct format.
First, a grouped barplot displays a numeric value (e.g. counts) split
in groups and subgroups. A few explanation about the code below: * the
input dataset must be a numeric matrix. Each group is a column. Each
subgroup is a row. So we can only deal with two variables at once. * the
barplot function will recognize this format, and
automatically perform the grouping for you. * the beside
option allows to toggle between the grouped and the stacked barchart
## make a table of counts by Treatment and Improved from the arthritis data
tab <- xtabs(~Improved+Treatment,data=arthritis)
tab## Treatment
## Improved Placebo Treated
## None 29 13
## Some 7 7
## Marked 7 21# Grouped barplot
barplot(tab, beside=TRUE, legend=rownames(tab), col=2:4)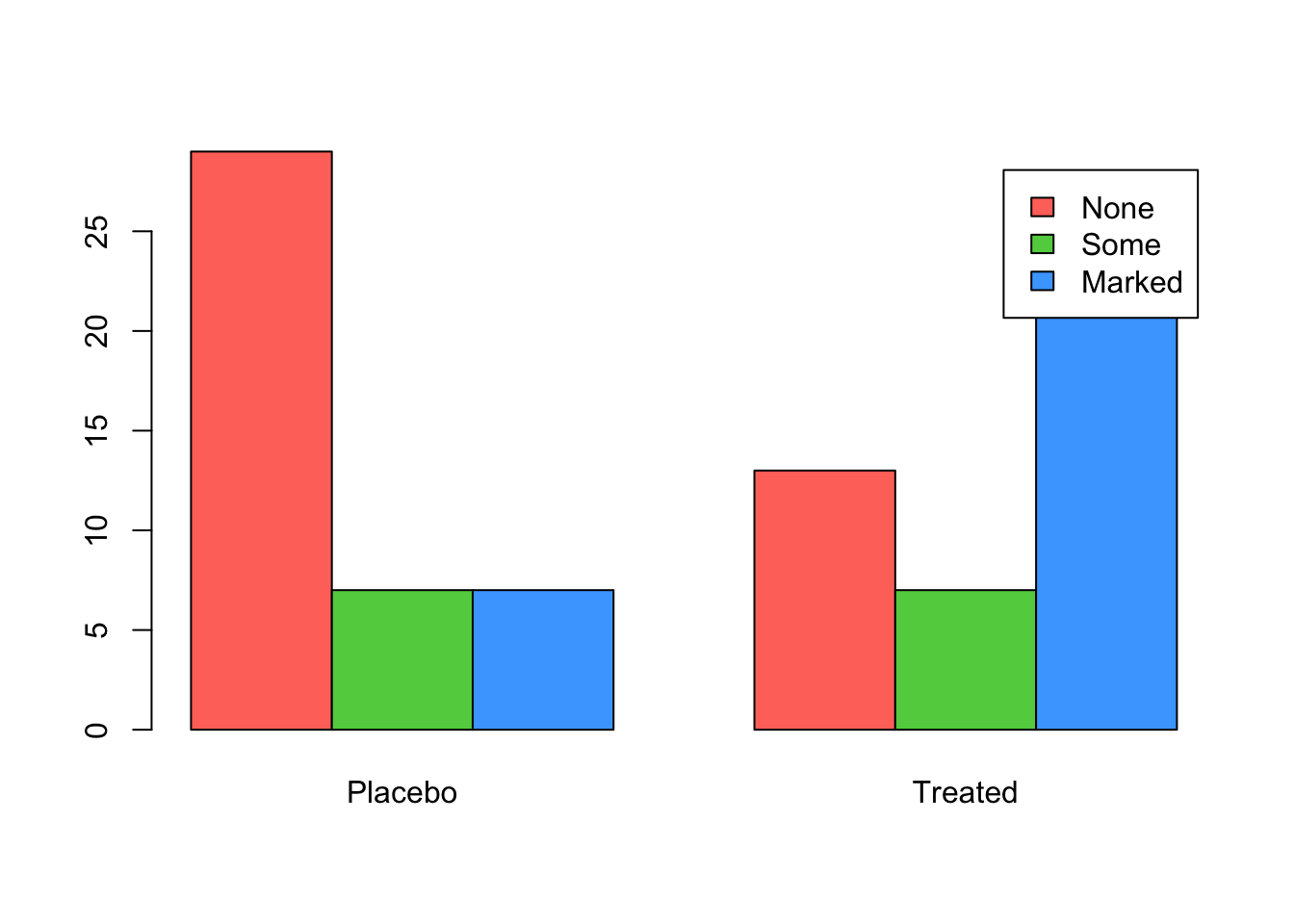
And to stack the bars, set beside=FALSE
barplot(tab, beside=FALSE, legend=rownames(tab), col=2:4)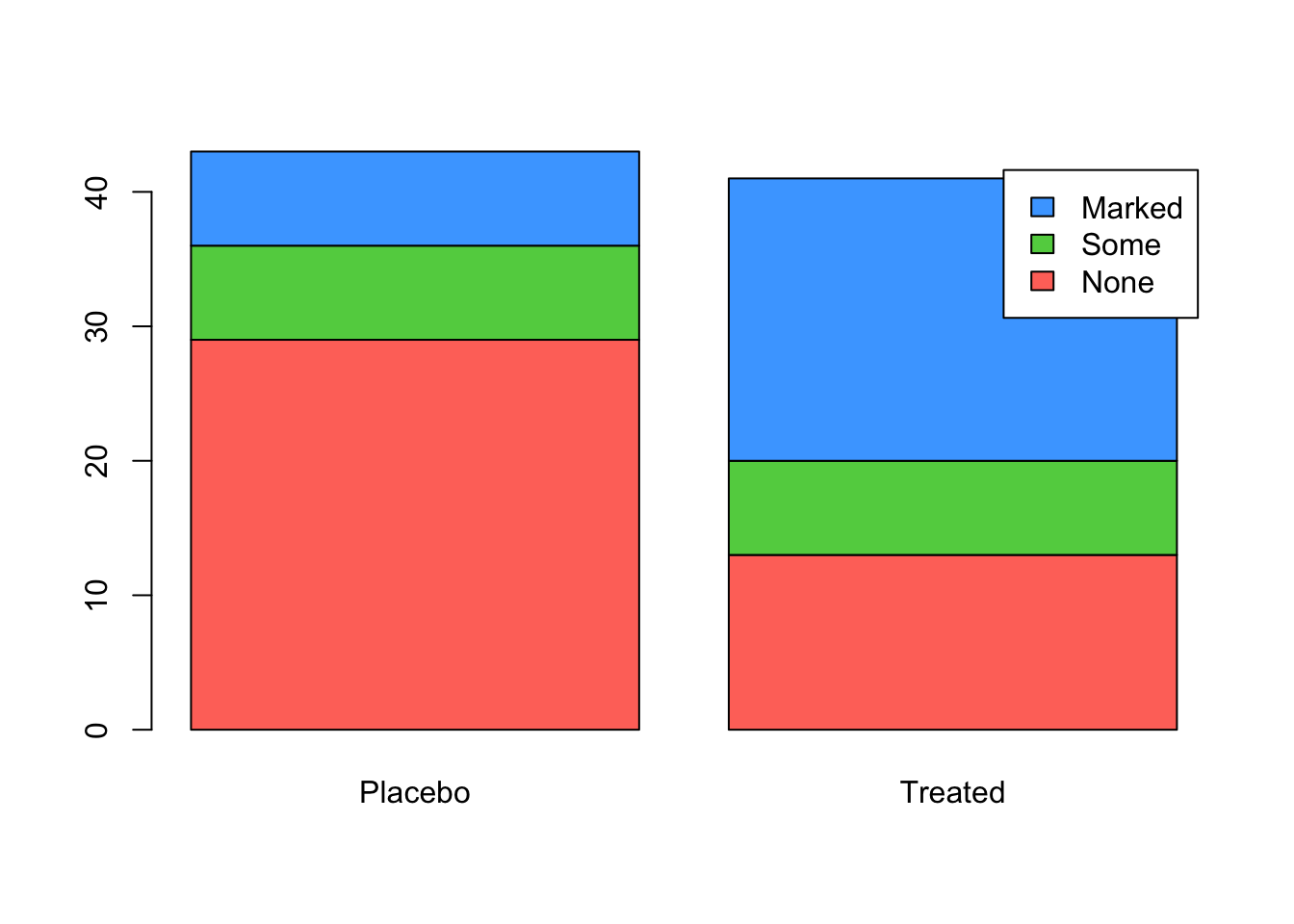
Stacked bars are often used to display the proportions of
the respective columns attributable to each sub-group. Thankfully, we
can easily convert tables of counts to proportions with the
prop.table function. If we want the proportions computed
within a column, set the margin=2 argument:
barplot(prop.table(tab, margin=2), beside=FALSE, legend=rownames(tab), col=2:4)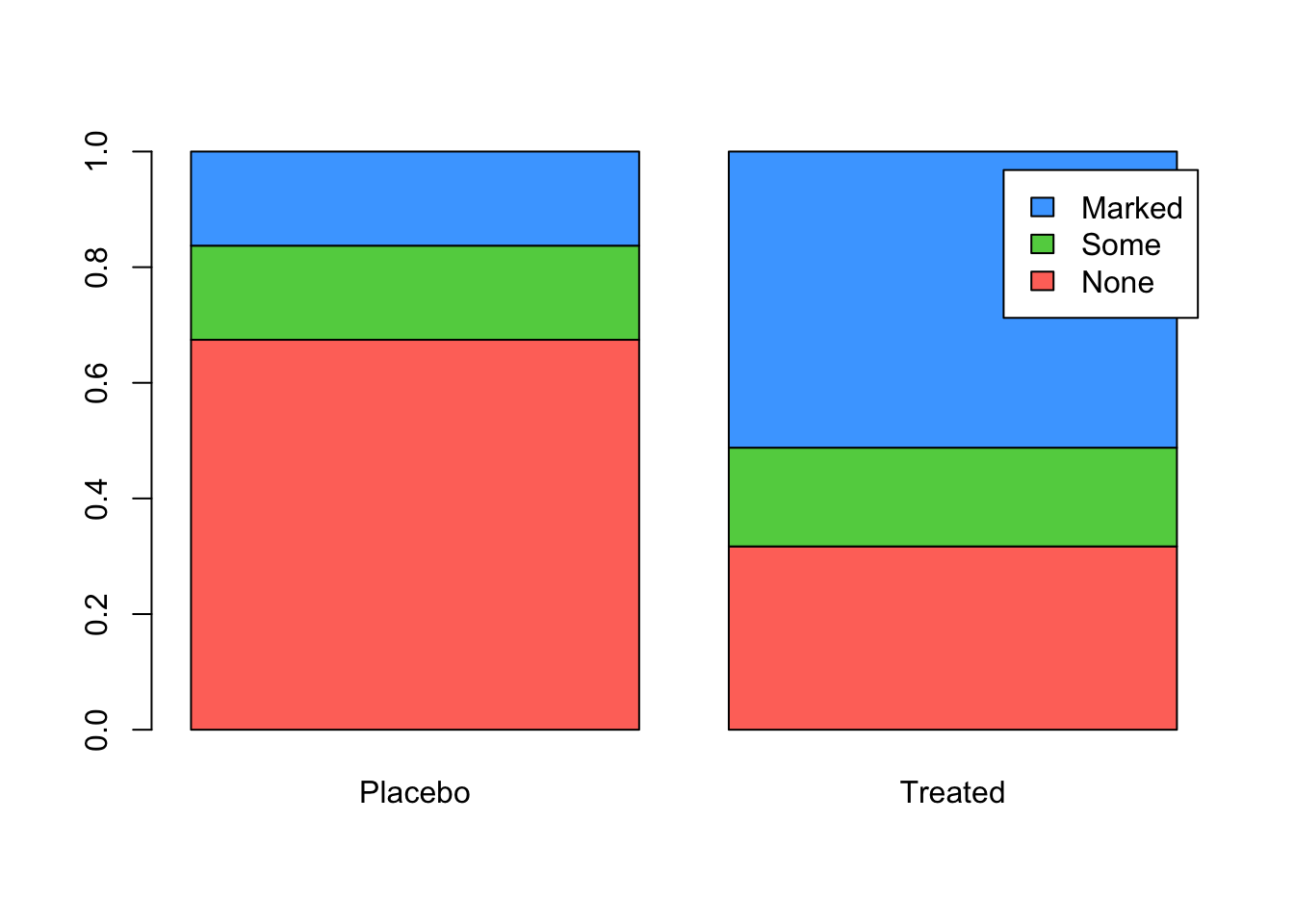
3 More Practice
3.1 Data set: Airline arrivals
Download data: airlineArrival
The airlineArrival data contains 11000 observations of 3
categorical variables: * Airport - a factor with levels
LosAngeles, Phoenix, SanDiego,
SanFrancisco, Seattle * Result -
a factor with levels Delayed,OnTime *
Airline - a factor with levels Alaska,
AmericaWest
- Is there much difference in the amount of delayed flights between the two airlines?
- What about the delays from different airports? Which look best for flights being on time? Which look worst?
- Now look at whether both Airport and Airline are associated with
delays. What do you find? Try turning on
shade=TRUE. - In which plot are the associations most pronounced?
3.2 Data set: Winter Olympic Medals
Download data: medals
The medals data contains the number of medals won at the
2016
Summer Olympics. The variables are * NOC - the country
* country - a factor indicating the country code *
medal - a factor with levels Bronze,
Silver, and Gold * count - the
number of medals of that type won by that country
- Try drawing a mosaic or doubledecker plot between
countryandmedal. - With the huge number of possible countries, this isn’t going to work.
- Let’s try some stacked barplots instead. Read the section on stacked barplots above.
- Make a stacked barplot showing the different countries on the x axis, with each bar split by the type of medal.
- There are probably too many countries here. Use the
Totalvariable to make a new data set containing only the records for countries which won more that 10 medals. Redraw your plot. - Try drawing the barplot horizontally (
horiz=TRUE) and rotate the labels (las=2). - Set the colours of the bars to use “#D4AF37” for Gold, “#C0C0C0” for Silver, and “#CD7F32” for Bronze.
- To finish off the plot, lets rearrange the bars into order
- Find the medal totals for each of the remaining countries, save this table to a variable.
- Apply the
colSumsfunction to the table to get the medal totals per country, and save this to another variable. - Use the
orderfunction to order the medal totals in decreasing order (decreasing=TRUE), and save this. - Now use the results of the
orderfunction at the previous step to rearrange the columns of the medals table, and save it. - Now draw the final barplot of our rearranged table!