R Techniques 10: Statistical methods
1 Statistical methods
1.1 Statistical tests in
the stats package
1.1.1 \(t\) tests
The t.test function produces a variety of t-tests.
Unlike most statistical packages, the default assumes unequal variance
of the two samples, however we can override this with the
var.equal argument.
# independent 2-sample t-test assuming equal variances and a pooled variance estimate
t.test(x,y,var.equal=TRUE)# independent 2-sample t-test unequal variance and the Welsh df modification.
t.test(x,y)
t.test(x,y,var.equal=FALSE)You can use the alternative="less" or
alternative="greater" options to specify a one-sided
test.
A paired-sample \(t\)-test is
obtained from the same function by specifying paired=TRUE
and ensuring both samples are the same size:
# paired t-test
t.test(x,y,paired=TRUE) Finally, a one-sample \(t\)-test can
be performed to test the population mean \(\mu\) of the sample against a specific
value by supplying only a single sample and a value for
mu:
# one sample t-test
t.test(y,mu=3) # Ho: mu=3R Help: t.test
1.1.2 Nonparametric tests of group differences
The nonparametric tests for comparing two samples can be performed in
much the same way, only this time we use the wilcox.test
function.
# independent 2-group Mann-Whitney U Test
wilcox.test(y,x) # where y and x are numeric# paired 2-group Signed Rank Test
wilcox.test(y1,y2,paired=TRUE) # where y1 and y2 are numericR Help: wilcox.test
1.2 Linear regression
1.2.1 Fitting linear
models with lm
Linear models can be fitted by the method of least squares in R using
the function lm. Suppose our response variable is \(y\), and we have a predictor variable \(x\) and we want to \(y\) as a linear function of \(x\), we can use lm to do
this:
model <- lm(y ~ x)Alternatively, if we have a data frame called dataset
with columns a and b then we could fit the
linear regression of a on b without having to
extract the columns by using the data argument
model <- lm(a ~ b, data=dataset)The function lm fits a linear model to data and we
specify the model using a ‘formula’ where the response variable is on
the left hand side separated by a ~ from the explanatory
variable(s). The formula provides a flexible way to specify various
different functional forms for the relationship. The data argument is
used to tell R where to look for the variables used in the formula.
Note: The ~ (tilde) symbol should be interpreted
as ‘is modelled as’.
For example, consider the data below from a study in central Florida where 15 alligators were captured and two measurements were made on each of the alligators. The weight (in pounds) was recorded with the snout vent length (in inches – this is the distance between the back of the head to the end of the nose). The data were analysed the data on the log scale (natural logarithms), and the goal is to determine whether there is a linear relationship between the variables:
alligator = data.frame(
lnLength = c(3.87, 3.61, 4.33, 3.43, 3.81, 3.83, 3.46, 3.76,
3.50, 3.58, 4.19, 3.78, 3.71, 3.73, 3.78),
lnWeight = c(4.87, 3.93, 6.46, 3.33, 4.38, 4.70, 3.50, 4.50,
3.58, 3.64, 5.90, 4.43, 4.38, 4.42, 4.25)
)
model <- lm(lnWeight~lnLength,data=alligator)Inspecting the fitted regression lm object shows us a
summary of the estimated model parameters:
model##
## Call:
## lm(formula = lnWeight ~ lnLength, data = alligator)
##
## Coefficients:
## (Intercept) lnLength
## -8.476 3.431We can also add a fitted regression line to a plot by simply passing
the fitted model to abline:
plot(y=alligator$lnWeight, x=alligator$lnLength)
abline(model,col='red')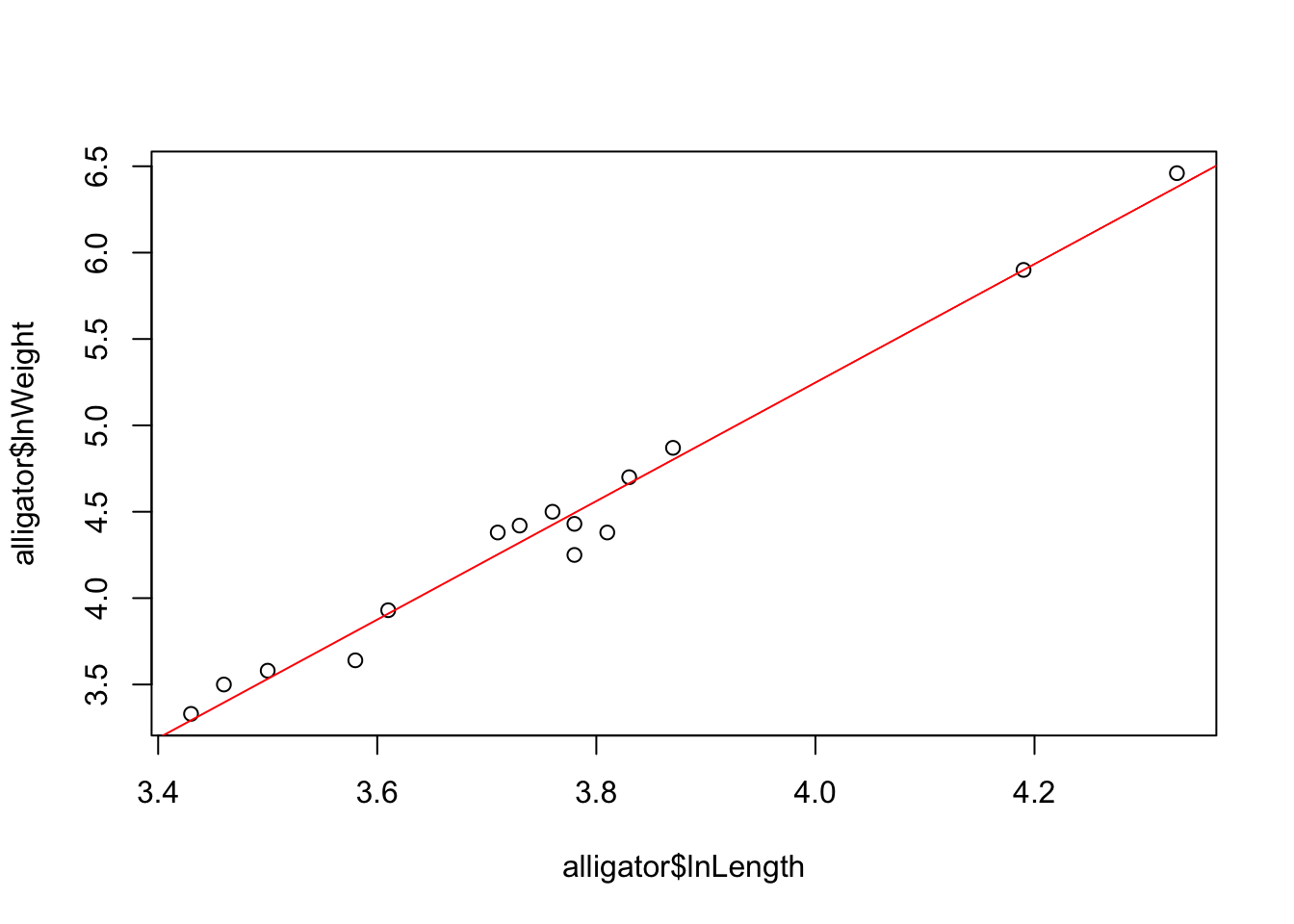
R Help: lm
1.2.2 The regression summary
The summary function in R is a multi-purpose function
that can be used to describe an R object. It can be applied to vectors,
matrices, data frames, linear models (lm objects), anova
decompositions and many other R objects. Applying summary to our
lm object via summary(model) will give a
summary of many key elements of the regrssion:
summary(model)##
## Call:
## lm(formula = lnWeight ~ lnLength, data = alligator)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.24348 -0.03186 0.03740 0.07727 0.12669
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -8.4761 0.5007 -16.93 3.08e-10 ***
## lnLength 3.4311 0.1330 25.80 1.49e-12 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1229 on 13 degrees of freedom
## Multiple R-squared: 0.9808, Adjusted R-squared: 0.9794
## F-statistic: 665.8 on 1 and 13 DF, p-value: 1.495e-12There is a lot of information in this output, but the key quantities are:
- Call: Printed here will be the formula of the regression model that was fitted.
- Residuals: This is a simple set of
summarystatistics of the residuals from the regression. Note that the mean of the residuals is always 0 and so is omitted. - Coefficients: This is a table containing
information about the fitted coefficients in the model and is the most
useful part of the output. Its columns are:
- The label of the fitted coefficient: The first will usually
be
(Intercept), indicating that the first row of the table contains information about the fitted intercept. Subsequent rows will be named after the other explanatory variables in the model forumula after the~ - The
Estimatecolumn gives the least squares estimates of the coefficients. - The
Std. Errorcolumn gives the corresponding standard error for each coefficient. - The
t valuecolumn contains the \(t\) test statistics for a test of the hypothesis \(H_0: \beta_i=0\) against \(H_0: \beta_i\neq 0\), for each coefficient \(\beta_i\). - The
Pr(>|t|)column is then the \(p\)-value associated with that each test, where a low p-value indicates that we have evidence to reject the null hypothesis that the estimate is different from 0 (with the significance levels given by the number of stars).
- The label of the fitted coefficient: The first will usually
be
- Residual standard error: This gives the square root of the unbiased estimate of the residual variance.
- Multiple R-Squared: For simple linear regression, this is the \(R^2\) value defined in lectures as the squared correlation coefficient and is a measure of goodness of fit (with values close to 1 indicating good fits to the data).
- Adjusted R-Squared: The adjusted R-Squared, \(\bar{R}^2\) is a penalised version of \(R^2\) which incorporates a penalty on the number of terms in the model to guard against unnecessarily overcomplicating the model. This is more useful when considering models with many predictors, and when comparing models with different numbers of predictors.
- F-statistic: and p-value: are the results of a hypothesis test that all coefficients other than the intercept are 0. This will be seen again in 3H.
R Help: summary
1.2.3 Extracting more detail
R also has a number of functions that, when applied to the results of a linear regression, will return key quantities such as residuals and fitted values.
coef(model)andcoefficicents(model)– returns the estimated model coefficients as a vector \((\widehat{\beta}_0,\widehat{\beta}_0)\)fitted(model)andfitted.values(model)– returns the vector of fitted values, \(\widehat{y}_i=\widehat{\beta}_0+\widehat{\beta}_0x_i\)resid(model)andresiduals(model)– returns the vector of residual values, \(e_i=y_i-\widehat{y}\)confint(model, level=0.95)– CIs for model parameterspredict(model, x, level=0.95)– predictions for the location of the SLR line, or for \(y\) at some new valuex.
We can also extract the individual elements from the summary output
from a regression analysis. Suppose we save the results of a call to the
summary function of a lm object as summ. By
using the $ we can extract even more valuable information
from the components of the regression summary:
summ$residuals– extracts the regression residuals asresidabovesumm$coefficients– returns the \(p \times 4\) coefficient summary table with columns for the estimated coefficient, its standard error, t-statistic and corresponding (two-sided) p-value.summ$sigma– the regression standard errorsumm$r.squared,summ$adj.r.squared– the regression \(R^2\) and adjusted \(R^2\) respectivelysumm$cov.unscaled– the \(p\times p\) (co)variance matrix for the least squares coefficients. The diagonal elements comprise the estimates of the variances for each of the \(p\) coefficients, and the off-diagonals give the estimated covariances between each pair of coefficients.
1.2.4 More complex linear models
The following commands are for future reference and show the syntax of how to create linear models with multiple predictors, models with quadratic (or higher power) terms, and models with interaction terms.
## multiple regression involving 2 predictors
model1 <- lm(y ~ x1 + x2, data=myData)
## quadratic regression on x1. Note each higher order term must be
## enclosed within the I() function
model2 <- lm(y ~ x1 + I(x1^2), data=myData)
## multiple regression with interaction
model3 <- lm(y ~ x1 + x2 + x1:x2, data=myData)
## more concise way of expressing model3
model3a <- lm(y ~ x1*x2, data=myData) R Help: lm, formula for more details on specifying a regression.